International Union of Pharmacology. XLI. Compendium of Voltage-Gated Ion Channels: Potassium Channels

Abstract
This summary article presents an overview of the molecular relationships among the voltage-gated potassium channels and a standard nomenclature for them, which is derived from the IUPHAR Compendium of Voltage-Gated Ion Channels.1 The complete Compendium, including data tables for each member of the potassium channel family can be found at http://www.iuphar-db.org/iuphar-ic/.
Almost a decade ago, a standardized nomenclature for the six-transmembrane domain (TM), voltage-gated K+ channel genes—the KV naming system—was widely adopted (Chandy et al., 1991; Gutman and Chandy, 1993). This nomenclature was based on deduced phylogenetic relationships; channels that shared 65% sequence identity being assigned to one subfamily. A parallel nomenclature—KCN—was developed by the Human Genome Organisation (HUGO) (White et al., 1997). Since then, the K+ channel superfamily of genes has greatly expanded, requiring an update of the naming system.
Structural Characteristics
Several structural classes of the potassium channels are now known, as illustrated by the transmembrane folding diagrams for this section of the Compendium, and most of these classes have multiple members.
The 6TM proteins include the KV channels, and the related small-conductance and intermediate-conductance Ca2+-activated K+ channels (KCa). Both the N and C termini of these proteins are located intracellularly, and the region between the fifth and sixth transmembrane domains (the P region) forms the ion conduction pathway. The functional channel is formed by the tetrameric association of these 6TM/1P subunits.
The second class of 2TM proteins includes the inward rectifiers, the KATP channels and the G protein-coupled channels (Doupnik et al., 1995). The N and C termini of these channels are also located cytoplasmically, the P region between the two transmembrane domains forms the pore, and the functional channel is a tetramer of these 2TM/1P subunits.
A third class has 7TM and encodes the large-conductance channel, Slo. There is a P region between TMVI and TMVII, and the channel functions as a tetramer, but unlike the other channels, Slo has its N terminus located extracellularly.
A fourth class of proteins has a 6TM/1P segment linked in tandem to a 2TM/1P segment, and the functional channel in this case is formed from the dimeric association of the 8TM/2P subunit.
Yet another class of channels, the K2P family, contains two 2TM/1P region-containing subunits linked in tandem, and the functional channel is a dimer of the 4TM/2P subunits.
Human Gene Nomenclature Committee System
The KCN system established by the Human Gene Nomenclature Committee (HGNC) of HUGO (Table 1) suffers from a lack of any rational basis for nomenclature and, in particular, ignores the structural and phylogenetic relationships of these proteins. For example, the KCNA–KCND subfamilies refer to 6TM/1P proteins, whereas the KCNE subfamily corresponds to an unrelated protein that functions as an accessory subunit to some families of K+ channels. Subfamilies encoding other 6TM/1P proteins include KCNF, KCNG, KCNH, KCNN, KCNQ, and KCNS, whereas KCNJ contains the 2TM/1P proteins, the KCNK subfamily corresponds to the 4TM/2P proteins, and the 7TM/1P is subfamily KCNMA1. Even though the Eag, Erg, and Elk channels are very different in their sequences, they are grouped together as KCNH; several members of this group, namely Elk1, Erg2, and Erg3, have not yet been assigned HGNC names. Similarly, the small-conductance (SKCa1–SKCa3) and intermediate-conductance (IKCa1) Ca2+-activated K+ channels are grouped together in the KCNN subfamily even though they share only ∼45% amino acid sequence identity and therefore are best considered as belonging to distinct subfamilies.
The KCN classification system
The Standardized K+ Channel Nomenclature System
The original KV nomenclature that created subfamilies KV1–KV6 has not been updated for several years. As new genes were discovered the name KV7 was skipped, and these new genes were instead assigned to subfamilies KV8 and KV9. Also, the more recently discovered KCNH (Eag, Erg, Elk) and KCNQ (KVLQT) subfamilies have not been included in the KV classification. A standardized nomenclature has been adopted in the case of the 2TM/1P channels, and subfamilies defined in this Kir family determined by the degree of sequence relatedness.
Formulation of a Rational Classification
With the completion of the mapping of the human genome, the time may be ripe for a re-evaluation of these issues, and the development of a uniform and rational nomenclature for all K+-selective channels. Such a naming system could incorporate the architectural similarities between different channel families, as well as their phylogenetic relationships. For example, the numerous 6TM/1P proteins and the sole 7TM/1P channel listed in Table 1 could be grouped into a single large family comprising multiple subfamilies defined according to their phylogenetic relatedness. Similarly, the 2TM/1P channels and the 2P channels could each be clustered into a separate family containing multiple subfamilies.
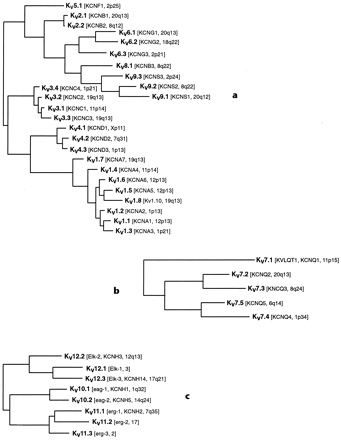
The standardized nomenclature for potassium channels presented in this compendium is shown in the phylogenetic trees of Figs. 1 through 4⇓⇓⇓. The 6TM/1P channels have been organized into two distinct groups based on their structural relatedness and predominant functional characteristics, namely the voltage-gated (KV) and calcium-activated (KCa) channels. The two-pore (K2P) and inward rectifier (Kir) channels likewise form two additional groups. Figure 1a shows the voltage-gated K+ channels of families KV1–KV6 and KV8–KV9, in a phylogenetic reconstruction using maximum parsimony based on an amino acid sequence alignment. Among this group, only KV1.8 currently lacks an HGNC name. The five members of a second KV group, the KV7 family (KCNQ1–KCNQ5), cannot readily be aligned with other KV channel proteins and are therefore shown in a separate tree in Fig. 1b. The three remaining KV families, KV10, KV11 and KV12, are closely enough related to each other to be shown in the single tree of Fig. 1c.
Phylogenetic trees of the KV channels. a, the KV1–KV6 and KV8–KV9 families; b, the KV7 family; c, the KV10–KV12 families. An amino acid sequence alignment made using CLUSTAL W was subjected to analysis by maximum parsimony (PAUP*). Only the hydrophobic core region of the alignment was used for analysis.
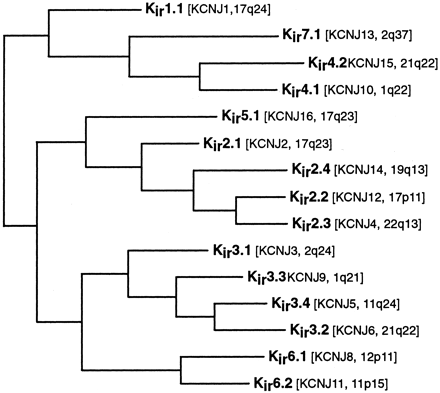
Phylogenetic tree showing the Kir families. See Fig. 1 for details of analysis.
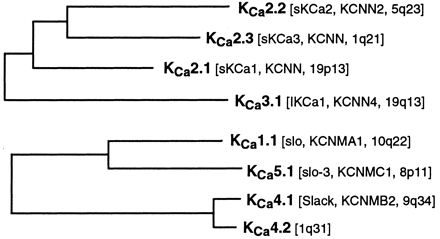
The five families comprising the calcium-sensitive potassium channels, KCa1–KCa5, numbered according to the order of their discovery, are shown in the two trees of Fig. 2. The phylogenetic relationship between the KCa1, KCa4, KCa5 group on the one hand, and the KCa2, KCa3 group on the other, is insufficiently clear at this time to readily connect them into a single tree.
Two phylogenetic trees showing the KCa1–KCa5 families. See Fig. 1 for details of analysis.
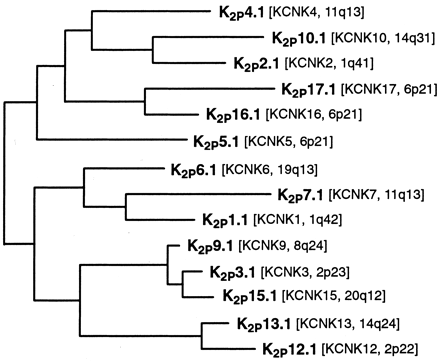
The two-pore or K2P potassium channels are shown in the tree of Fig. 3. The numbers have been taken from the HGNC “KCNK” numbering, without resequencing around the missing numbers (namely 8, 11, and 14), and without consolidating clearly related channels into subfamilies. A consensus among researchers in the field will hopefully establish a more rational nomenclature in the future.
Phylogenetic tree showing the K2P families. See Fig. 1 for details of analysis.
The last remaining group of potassium channels, the inward rectifier or Kir channels, is represented in the tree of Fig. 4, using the previously established nomenclature and subfamily groupings.
Footnotes
-
↵1 This work was previously published in Catterall WA, Chandy KG, and Gutman GA, eds. (2002) The IUPHAR Compendium of Voltage-Gated Ion Channels, International Union of Pharmacology Media, Leeds, UK.
-
DOI: 10.1124/pr.55.4.9.
- The American Society for Pharmacology and Experimental Therapeutics