Abstract
ERBB4 (HER4) is a member of the ERBB family of receptor tyrosine kinases, a family that includes the epidermal growth factor receptor (EGFR/ERBB1/HER1), ERBB2 (Neu/HER2), and ERBB3 (HER3). EGFR and ERBB2 are oncoproteins and validated targets for therapeutic intervention in a variety of solid tumors. In contrast, the role that ERBB4 plays in human malignancies is ambiguous. Thus, here we review the literature regarding ERBB4 function in human malignancies. We review the mechanisms of ERBB4 signaling with an emphasis on mechanisms of signaling specificity. In the context of this signaling specificity, we discuss the hypothesis that ERBB4 appears to function as a tumor suppressor protein and as an oncoprotein. Next, we review the literature that describes the role of ERBB4 in tumors of the bladder, liver, prostate, brain, colon, stomach, lung, bone, ovary, thyroid, hematopoietic tissues, pancreas, breast, skin, head, and neck. Whenever possible, we discuss the possibility that ERBB4 mutants function as biomarkers in these tumors. Finally, we discuss the potential roles of ERBB4 mutants in the staging of human tumors and how ERBB4 function may dictate the treatment of human tumors.
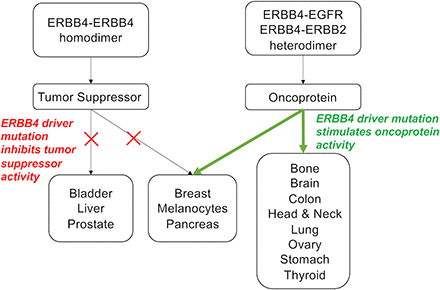
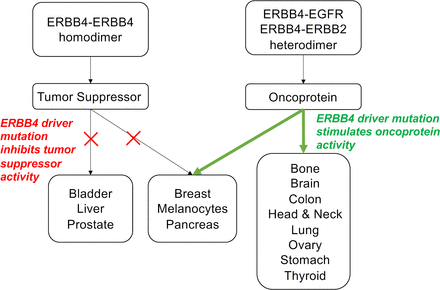
Significance Statement This articles reviews ERBB4 function in the context of the mechanistic model that ERBB4 homodimers function as tumor suppressors, whereas ERBB4-EGFR or ERBB4-ERBB2 heterodimers act as oncogenes. Thus, this review serves as a mechanistic framework for clinicians and scientists to consider the role of ERBB4 and ERBB4 mutants in staging and treating human tumors.
I. Introduction
ERBB4 (HER4) is a member of the ERBB family of receptor tyrosine kinases, a family that includes the epidermal growth factor receptor (EGFR/ERBB1/HER1), ERBB2 (Neu/HER2), and ERBB3 (HER3) (Carraway et al., 1997; Gullick and Srinivasan, 1998; Riese and Stern, 1998; Olayioye et al., 2000; Schlessinger, 2000; Yarden and Sliwkowski, 2001; Burgess et al., 2003; Earp et al., 2003; Holbro and Hynes, 2004; Zaczek et al., 2005; Citri and Yarden, 2006; Britsch, 2007; Karamouzis et al., 2007; Riese et al., 2007; Burgess, 2008; Mei and Xiong, 2008; Bose and Zhang, 2009; Lemmon, 2009; Wilson et al., 2009; Lemmon and Schlessinger, 2010; Rudloff and Samuels, 2010; Easty et al., 2011; Eccles, 2011; Arteaga and Engelman, 2014; Bessman et al., 2014; Lemmon et al., 2014; Riese and Cullum, 2014; Roskoski, 2014; Alaoui-Jamali et al., 2015; Appert-Collin et al., 2015; Kennedy et al., 2016; Mishra et al., 2017; Wang, 2017). EGFR and ERBB2 are extensively studied oncoproteins and are well validated targets for therapeutic intervention in various solid tumors. ERBB3 also appears to function as an oncoprotein, although its role in human malignancies appears to be more limited than the roles of EGFR or ERBB2. In contrast, the role that ERBB4 plays in human malignancies is ambiguous (Klijn et al., 1992; Lupu et al., 1992; Tripathy and Benz, 1992; Hynes and Stern, 1994; Lupu et al., 1995; Carraway et al., 1997; Gullick and Srinivasan, 1998; Riese and Stern, 1998; Olayioye et al., 2000; Schlessinger, 2000; Bowers et al., 2001; Hynes et al., 2001; Yarden and Sliwkowski, 2001; Anderson and Ahmad, 2002; Zhou and Carpenter, 2002; Carpenter, 2003a; Earp et al., 2003; Holbro and Hynes, 2004; Roskoski, 2004; Hynes and Lane, 2005; Zaczek et al., 2005; Citri and Yarden, 2006; Engelman and Cantley, 2006; Hynes and Schlange, 2006; Nicholas et al., 2006; Schlessinger and Lemmon, 2006; Breuleux, 2007; Karamouzis et al., 2007; Riese et al., 2007; Burgess, 2008; Black and Dinney, 2008; Chuu et al., 2008; Jones, 2008; Lafky et al., 2008; Muraoka-Cook et al., 2008; Paatero and Elenius, 2008; Roepstorff et al., 2008; Sorkin and Goh, 2008; Stern, 2008; Uberall et al., 2008; Lemmon, 2009; Wilson et al., 2009; Carraway, 2010; Hollmen and Elenius, 2010; Koutras et al., 2010; Lemmon and Schlessinger, 2010; Rudloff and Samuels, 2010; Easty et al., 2011; Eccles, 2011; Veikkolainen et al., 2011; Lindet et al., 2012; Khelwatty et al., 2013; Arteaga and Engelman, 2014; Gala and Chandarlapaty, 2014; Ma et al., 2014; Modjtahedi et al., 2014; Appert-Collin et al., 2015; Feldinger and Kong, 2015; Cao et al., 2016; Kennedy et al., 2016; Lemmon et al., 2016; Sacco and Worden, 2016; Wang et al., 2016; Kourie et al., 2017; Mishra et al., 2017; Mota et al., 2017; Lyu et al., 2018; Arienti et al., 2019; Black et al., 2019; Jordan et al., 2019; Maennling et al., 2019; Wang et al., 2019; Segers et al., 2020). Some data indicate that ERBB4 functions as an oncoprotein, making it a potential target for therapeutic intervention. Other data indicate that ERBB4 functions as a tumor suppressor and is therefore unsuited for direct therapeutic intervention.
Genome-wide, next-generation sequencing of tumor cell transcripts has identified mutations and changes in gene expression that are characteristics of tumor genesis and progression. This has enabled a revolution in mechanism-based targeted chemotherapy. However, the absence of a clear-cut understanding of the role that ERBB4 plays in human malignancies has hindered the translation of ERBB4 expression and ERBB4 mutation data into clinical practice.
Hence, here we will review ERBB4 function, including mechanisms by which biochemical and biologic responses to ERBB4 signaling are specified. We will emphasize the mechanistic model that ERBB4 homodimerization and heterodimerization can be coupled to distinct biochemical responses, thereby enabling ERBB4 homodimers to function as tumor suppressors and ERBB4 heterodimers to function as oncoproteins. We will review past efforts to characterize the role of ERBB4 in human malignancies in the context of this mechanistic model, and we will discuss the implications of this model in the staging and treatment of human tumors.
II. Structure and Function of ERBB4
A. Functional Domains of ERBB Receptors
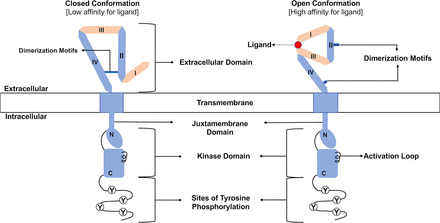
The four ERBB family receptors share structural homology (Fig. 1). They each contain a large, amino-terminal, extracellular region comprised of four subdomains (I–IV) involved in ligand binding and receptor dimerization. Adjacent to the extracellular domain is a single-pass, hydrophobic transmembrane domain. Carboxyl terminal to the transmembrane domain is a cytoplasmic tyrosine kinase domain and a tail region that features numerous tyrosine residues that serve as sites of phosphorylation (Carraway et al., 1997; Gullick and Srinivasan, 1998; Riese and Stern, 1998; Olayioye et al., 2000; Hynes et al., 2001; Burgess et al., 2003; Holbro and Hynes, 2004; Roskoski, 2004, 2014; Britsch, 2007; Burgess, 2008; Lemmon et al., 2014; Alaoui-Jamali et al., 2015; Appert-Collin et al., 2015; Wang, 2017; Maennling et al., 2019).
Organization of ERBB receptors. The amino-terminal extracellular region consists of four subdomains (I–IV) responsible for ligand binding and receptor dimerization. Note the dimerization motifs in subdomains II and IV that stabilize the intramolecular interactions characteristic of the closed conformation and enable the intermolecular interactions necessary for dimerization of two receptor molecules that exist in the open conformation. A hydrophobic transmembrane domain lies between the extracellular region and the cytoplasmic tyrosine kinase domain. This kinase domain can be divided into amino-terminal (N) and carboxyl-terminal (C) lobes. Several sites of tyrosine phosphorylation (Y) reside at the carboxyl terminus of these receptors. Finally, note that ligand binding stabilizes a receptor molecule in the open conformation.
B. General Principles of Ligand-Induced ERBB Receptor Dimerization
The ligands for ERBB receptors are members of the epidermal growth factor (EGF) family of peptide growth factors. No ligand has been identified for ERBB2, whereas multiple EGF family members (discussed in Section II.F) bind to EGFR, ERBB3, and ERBB4 (Fig. 2). In the absence of a cognate ligand, most EGFR, ERBB3, and ERBB4 molecules on the cell surface exist in a closed conformation that features intramolecular interactions between extracellular subdomains (ECDs) II and IV (Fig. 1). This conformation buries the receptor dimerization motifs found in ECDs II and IV and increases the distance between the ligand-binding motifs found in ECDs I and III. This spatial distance prevents ligand molecules from simultaneously binding the ligand-binding motifs found in ECDs I and III. Thus, in the closed conformational state, EGFR, ERBB3, and ERBB4 bind ligands with low affinity and fail to dimerize and signal.
Ligand-induced ERBB receptor signaling. Eleven members of the EGF family of peptide growth factors bind to EGFR, ERBB3, and ERBB4. Some EGF family members bind to multiple ERBB family receptors, but no EGF family member binds to ERBB2. The binding of an EGF family hormone to a receptor stabilizes the extracellular domain of the receptor in an open conformation that enables receptor dimerization. ERBB receptors can either homodimerize or heterodimerize. The symmetrical dimerization of the extracellular region of two ERBB receptors causes asymmetric dimerization of the cytoplasmic regions. This asymmetric dimerization enables the kinase domain of one monomer to allosterically stimulate the kinase activity of the other monomer. This results in transphosphorylation of one monomer by the other on tyrosine residues.
In the absence of ligand, a small fraction of EGFR, ERBB3, and ERBB4 molecules on the cell surface exist in an open conformation that lacks the interactions between ECDs II and IV. This conformation exposes the receptor dimerization motifs found in ECDs II and IV. Therefore, this open conformation permits ligand-independent receptor dimerization and signaling, particularly during extreme receptor overexpression (≥10⋀6 receptor molecules/cell); this overexpression enables stochastic dimerization of two receptor molecules in the open conformation (note that extreme receptor overexpression is observed in some types of human tumors, as will be discussed in later sections). However, to reiterate, EGFR, ERBB3, and ERBB4 molecules exist in an equilibrium between the closed and open conformations that strongly favors the closed conformation, thereby limiting ligand-independent receptor signaling under physiologic levels of expression.
When EGFR, ERBB3, or ERBB4 molecules are in the open conformation, there is a relatively small distance between ECDs I and III, such that it permits simultaneous, high-affinity binding of a single-ligand molecule to the ligand-binding sites of both subdomains (Fig. 1). Ligand binding stabilizes the receptor molecule in the open conformation, thereby facilitating receptor dimerization and signaling (Fig. 2) (Lemmon et al., 1997, 2014; Riese and Stern, 1998; Cho and Leahy, 2002; Garrett et al., 2002; Burgess et al., 2003; Ferguson et al., 2003, 2020; Holbro and Hynes, 2004; Roskoski, 2004, 2014; Bouyain et al., 2005; Dawson et al., 2005; Citri and Yarden, 2006; Ozcan et al., 2006; Britsch, 2007; Riese et al., 2007; Lemmon, 2009; Lemmon and Schlessinger, 2010; Rudloff and Samuels, 2010; Alaoui-Jamali et al., 2015; Appert-Collin et al., 2015; Feldinger and Kong, 2015; Kennedy et al., 2016; Wang, 2017; Maennling et al., 2019).
The effects of ligand binding on ERBB4 dimerization and signaling have been modeled by the constitutive dimerization and phosphorylation of the synthetic ERBB4 Q646C (Penington et al., 2002) and I658E (Vidal et al., 2007) mutants. The Q646C mutant, which undergoes homodimerization but not heterodimerization with other ERBB receptors, is functionally distinct from constitutively active ERBB2 mutants in that it does not cause malignant growth transformation of fibroblasts. Instead, the Q646C mutant inhibits clonogenic proliferation by various human breast, prostate, and pancreatic tumor cell lines (Penington et al., 2002; Williams et al., 2003; Gallo et al., 2006, 2013; Pitfield et al., 2006; Mill et al., 2011a). Likewise, the I658E mutant promotes apoptosis in numerous human breast, prostate, and ovarian tumor cell lines (Vidal et al., 2007). Consequently, these mutants indicate that ERBB4 homodimers function as tumor suppressors in a variety of contexts. We will discuss this general principle in more detail throughout this review.
C. General Principles of ERBB Receptor Tyrosine Phosphorylation
The dimerization of the extracellular subdomains of two ERBB receptor molecules causes dimerization of the intracellular subdomains of those molecules (Fig. 2). The extracellular domains of two receptor molecules form a symmetrical dimer. In contrast, the intracellular domains form an asymmetric dimer in which one monomer (the “regulatory/substrate monomer”) causes a conformational change in the other monomer (the “catalytic monomer”). This change in conformation activates the tyrosine kinase activity of the catalytic monomer, resulting in phosphorylation of tyrosine residues of the regulatory/substrate monomer. The carboxyl terminus of each ERBB receptor molecule contains multiple tyrosine residues, and this region is relatively unstructured. Thus, ERBB4 receptor dimerization and signaling features phosphorylation of multiple tyrosine residues within a population of receptor molecules (Fig. 3) (Carraway and Cantley, 1994; Jeffrey et al., 1995; Carraway et al., 1997; Olayioye et al., 2000; Bowers et al., 2001; Yarden and Sliwkowski, 2001; Landau et al., 2004; Jones et al., 2006; Zhang et al., 2006; Britsch, 2007; Qiu et al., 2008; Bose and Zhang, 2009; Foley et al., 2010; Lemmon and Schlessinger, 2010; Lemmon et al., 2014; Riese and Cullum, 2014; Roskoski, 2014; Alaoui-Jamali et al., 2015; Kovacs et al., 2015; Kennedy et al., 2016; Mishra et al., 2017; Wang, 2017; Black et al., 2019; Segers et al., 2020).
The cytoplasmic domain of ERBB4 possesses several candidate and validated sites of tyrosine phosphorylation. These sites of ERBB4 tyrosine phosphorylation are depicted along with candidate and validated effector proteins that directly or indirectly interact with these phosphorylation sites. The cytoplasmic domain is not depicted to scale.
D. Canonical ERBB4 Signaling Relies on ERBB4 Tyrosine Phosphorylation
The phosphorylation of ERBB receptors creates binding sites for numerous cytoplasmic signaling proteins that possess SRC homology domain 2 or (to a lesser extent) phosphotyrosine binding domain domains. The binding of these signaling proteins can trigger numerous intracellular signaling cascades, resulting in a wide range of phenotypes. The recognition of a specific site of tyrosine phosphorylation of an ERBB receptor by a specific cytoplasmic protein that possesses an SRC homology domain 2 or phosphotyrosine binding domain is dependent in part on the ERBB receptor amino-acid residues immediately adjacent to the phosphorylated tyrosine residue. Therefore, different sites of tyrosine phosphorylation can couple to different cytoplasmic signaling effectors, resulting in distinct phenotypes (Fig. 3) (Carraway and Cantley, 1994; Carraway et al., 1997; Olayioye et al., 2000; Yarden and Sliwkowski, 2001; Schlessinger and Lemmon, 2003; Holbro and Hynes, 2004; Schulze et al., 2005; Zaczek et al., 2005; Citri and Yarden, 2006; Jones et al., 2006; Kaushansky et al., 2008; Lemmon and Schlessinger, 2010; Eccles, 2011; Riese and Cullum, 2014; Roskoski, 2014; Kovacs et al., 2015; Wang, 2017; Black et al., 2019; Segers et al., 2020).
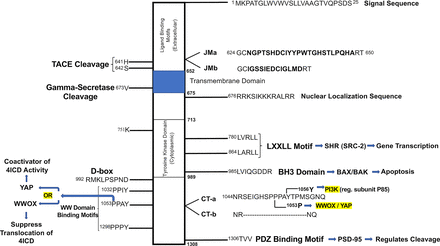
Mass spectrometry, high-performance liquid chromatography, and other analytical approaches have identified candidate sites of ERBB4 tyrosine phosphorylation. Protein microarrays and other methods have identified cytoplasmic signaling proteins that bind to 21 of these sites of phosphorylation (Fig. 3) (Schulze et al., 2005; Kaushansky et al., 2008; Segers et al., 2020). Additional putative ERBB4 signaling modulators and effectors that appear to bind the ERBB4 cytoplasmic domain independent of specific sites of tyrosine phosphorylation have been identified (Fig. 4). Altogether, an incomplete list of the proteins that apparently mediate ERBB4 function via physical and functional interactions with the ERBB4 cytoplasmic domain includes the Rous sarcoma virus protein tyrosine kinase (SRC); the Abelson-related protein tyrosine kinase (ABL2); the spleen-associated protein tyrosine kinase (SYK); the Janus kinase 1 (JAK1); protein tyrosine phosphatase non-receptor type 11 (PTPN11); phospholipase C gamma-2 (PLCG2); phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit alpha isoform (PIK3CA) via its p85 regulatory subunit; the Casitas B-lineage lymphoma ubiquitin ligase (CBL); the second guanine nucleotide exchange factor named after the sixth letter of the Greek alphabet (VAV2); the rat sarcoma virus protein (RAS) GTPase activating protein RASA1; the signal transducer and activator transcription factors STAT1 and STAT5a; the Yes-associated protein 1 (YAP1); the tumor suppressor oxidoreductase WWOX; postsynpatic density protein 95 (PSD-95), which binds the PDZ-domain recognition motif at the carboxyl terminus of ERBB4; and several adapter proteins, including the SRC homology domain 2-containing transforming protein (SHC1), growth factor receptor-bound protein 2 (GRB2), growth factor receptor-bound protein 7 (GRB7), CT10 regulator of kinase (CRK), and the CRK-like protein (CRKL) (Culouscou et al., 1995; Sepp-Lorenzino et al., 1996; Elenius et al., 1997b; Fiddes et al., 1998; Olayioye et al., 1998; Pinkas-Kramarski et al., 1998b; Wang et al., 1998; Elenius et al., 1999; Jones et al., 1999; Olayioye et al., 1999; Garcia et al., 2000; Huang et al., 2000, 2002; Sweeney et al., 2000; Puricelli et al., 2002; Carpenter, 2003a,b; Schulze et al., 2005; Chuu et al., 2008; Kaushansky et al., 2008; Ishibashi et al., 2013; Roskoski, 2014; Li et al., 2015; Wang, 2017; Segers et al., 2020). Section II.H, which is focused on signaling by the soluble ERBB4 cytoplasmic domain (4ICD), further discusses ERBB4 signaling effectors, some of which are not mentioned in Section II.D.
Additional motifs and effectors that modulate and mediate ERBB4 signaling. The cytoplasmic region of ERBB4 is depicted along with the differences in the amino-acid sequence of the four ERBB4 splicing isoforms. Sites of ERBB4 functional motifs are indicated, along with the particular downstream signaling effectors that bind to these sites. The cytoplasmic region is not depicted to scale.
E. Differences in Phosphorylation Sites Enable ERBB Receptor Signaling Specificity
EGFR and ERBB2 do not possess a phosphorylation site that enables direct binding of the p85 regulatory subunit of phosphatidylinositol 3-kinase (PI3K); in contrast, both ERBB3 and ERBB4 do possess at least one phosphorylation site that enables direct binding of p85 and activation of PI3K signaling (Yarden and Sliwkowski, 2001; Citri and Yarden, 2006; Arteaga and Engelman, 2014). Similarly, an EGFR phosphorylation site binds to the ubiquitin ligase CBL, resulting in CBL-dependent EGFR downregulation. In contrast, ERBB4 does not appear to directly bind CBL but instead requires the adaptor protein GRB2 for CBL binding (Carraway and Sweeney, 2001; Citri and Yarden, 2006; Carraway, 2010; Kiuchi et al., 2014; Roskoski, 2014). These differences in coupling to signaling effectors may account for some of the specificity in receptor coupling to biochemical pathways and biologic responses. For example, signaling by EGFR or ERBB2 homodimers typically causes increased cell survival or proliferation, whereas signaling by ERBB4 homodimers typically results in tumor suppression (Riese et al., 1996a,b; Muthuswamy et al., 1999; Sartor et al., 2001; Penington et al., 2002; Earp et al., 2003).
F. Differences in Ligand Potency and Intrinsic Activity Enable ERBB Receptor Signaling Specificity
The EGF family of peptide growth factors (Fig. 2) consists of 11 family members, each encoded by a distinct gene: amphiregulin (AREG), betacellulin (BTC), EGF, epigen, epiregulin (EREG), heparin-binding EGF-like growth factor (HBEGF), neuregulin1 (NRG1), neuregulin2 (NRG2), neuregulin3 (NRG3), neuregulin4 (NRG4), and transforming growth factor α (TGFA). Moreover, there are multiple, functionally distinct splicing isoforms of both neuregulin1 and neuregulin2. Several of these ligands exhibit differences in receptor binding affinity. For example, NRG1 binds to ERBB3 and ERBB4; EREG, HBEGF, and BTC bind to EGFR and ERBB4; and NRG3 and NRG4 bind only to ERBB4 (Fig. 2) (Alimandi et al., 1997; Pinkas-Kramarski et al., 1998a; Olayioye et al., 2000; Hynes et al., 2001; Carpenter, 2003a; Zaczek et al., 2005; Karamouzis et al., 2007; Wilson et al., 2009; Eccles, 2011; Veikkolainen et al., 2011; Arteaga and Engelman, 2014; Dahlhoff et al., 2014; Macdonald-Obermann and Pike, 2014; Riese and Cullum, 2014; Roskoski, 2014; Feldinger and Kong, 2015; Arienti et al., 2019; Black et al., 2019; Maennling et al., 2019; Segers et al., 2020).
Moreover, saturating concentrations of different ligands at the same receptor may elicit distinct effects (Wilson et al., 2009). For example, a saturating concentration of EREG, AREG, or TGFA stimulates greater EGFR coupling to cell proliferation than a saturating concentration of EGF. This difference appears to reflect that, unlike AREG, EGF stimulates EGFR phosphorylation at Tyr1045, a canonical binding site for the E3 ubiquitin protein ligase CBL; thus, compared with AREG, EGF causes greater EGFR ubiquitination and downregulation and diminished EGFR signaling duration. Consistent with this model, the EGFR Y1045F mutant causes EGF to exhibit greater intrinsic activity but does not affect the intrinsic activity of AREG. Indeed, in the presence of the EGFR Y1045F mutant, EGF and AREG exhibit roughly equal intrinsic activity (Levkowitz et al., 1998; Shelly et al., 1998; Komurasaki et al., 2002; Joslin et al., 2007; Gilmore et al., 2008, 2009; Stern et al., 2008; Jansen et al., 2009; Roepstorff et al., 2009; Willmarth et al., 2009; Wilson et al., 2009; Ahsan et al., 2010; Foley et al., 2010; Wilson et al., 2012a; Macdonald-Obermann and Pike, 2014; Riese and Cullum, 2014; Lemmon et al., 2016; Wee and Wang, 2018).
As noted elsewhere, subtle differences in the structure of receptor monomers within a receptor dimer or subtle differences in the juxtapositioning of receptor monomers within a receptor dimer appear to determine which receptor tyrosine residues become phosphorylated, how receptors are processed after ligand engagement, the identity and duration of downstream signaling events, and which biologic responses result from these actions. Consistent with this model, EGF and TGFA induce distinct conformations of the EGFR intracellular region (Wilson et al., 2009; Foley et al., 2010; Scheck et al., 2012; Bessman et al., 2014). Similarly, EREG and EGF induce distinct conformations of the EGFR extracellular region. Moreover, EREG induces less stable EGFR dimers than does EGF. These differences may account for the failure of EREG to stimulate EGFR phosphorylation at Tyr1045 and the fact that EREG stimulates EGFR signaling of greater duration than does EGF (Freed et al., 2017).
Noteworthy differences in ligand efficacy can also be observed among the ERBB4 ligands (Wilson et al., 2009). NRG2 encodes two distinct sets of splicing isoforms that display substantial differences in the amino-acid sequence of the EGF homology domain, which is responsible for receptor binding. The isoform NRG2β binds to ERBB3 and ERBB4 with relatively high affinity, potently stimulates phosphorylation of both receptors (the former in the context of an ERBB3-ERBB2 heterodimer), and stimulates coupling of ERBB3-ERBB2 heterodimers or ERBB4-EGFR heterodimers to cell survival and proliferation. In contrast, NRG2α binds to ERBB3 and ERBB4 with relatively low affinity and stimulates phosphorylation of both receptors with relatively low potency. Moreover, a saturating concentration of NRG2α fails to stimulate the coupling of ERBB3-ERBB2 heterodimers or ERBB4-EGFR heterodimers to cell proliferation. The failure of NRG2α to stimulate the coupling of ERBB3-ERBB2 heterodimers and ERBB4-EGFR heterodimers to cell proliferation is not due to a failure of NRG2α to bind to ERBB3 or ERBB4, respectively. Indeed, NRG2α competitively antagonizes the action of NRG2β at either ERBB3 or ERBB4 (Hobbs et al., 2002; Cote et al., 2005; Gilmore et al., 2006; Wilson et al., 2007, 2009, 2012b; Eckert et al., 2009).
Consistent with the current mechanistic explanation for differences in EGFR ligand efficacy, it has been postulated that NRG2α and NRG2β cause ERBB4 to dimerize in slightly different conformations, resulting in the phosphorylation of different sets of tyrosine residues and differential coupling to downstream signaling effectors (Sweeney et al., 2000; Wilson et al., 2009). This mechanistic model is consistent with the observation that NRG2β stimulates ERBB4-EGFR heterodimers to couple to PI3K signaling via phosphorylation of ERBB4 Tyr1056 by EGFR; in contrast, NRG2α fails to stimulate PI3K signaling, presumably due to a failure of NRG2α to stimulate ERBB4 phosphorylation at Tyr1056 (Cote et al., 2005; Eckert et al., 2009; Wilson et al., 2012b). Moreover, this mechanistic model is consistent with the observation that altering the juxtapositioning of ERBB2 monomers within an ERBB2 homodimer affects the ability of these homodimers to stimulate malignant growth transformation (Burke and Stern, 1998; Wilson et al., 2009); likewise, this mechanistic model is consistent with the observation that altering the juxtapositioning of ERBB4 monomers within an ERBB4 homodimer affects the ability of these homodimers to function as tumor suppressors (Williams et al., 2003; Gallo et al., 2006; Pitfield et al., 2006; Wilson et al., 2009).
G. ERBB Receptor Homodimers and Heterodimers Enable Signaling Specificity
As noted earlier, ligand binding and stabilization of the receptor extracellular domain in the open conformation enables dimerization with another receptor molecule in the open conformation. Under many conditions, homodimerization predominates as the presence of a ligand increases the availability of its cognate receptor in the open conformation. However, heterodimerization of a liganded receptor to an unliganded receptor can occur, particularly when the unliganded receptor is overexpressed; overexpression increases the concentration of the unliganded receptor in the open conformation. Similarly, because the structure of the ERBB2 extracellular domain resembles the open (ligand-bound) conformation of the EGFR, ERBB3, and ERBB4 extracellular domains (Fig. 2), ERBB2 is a preferred heterodimerization partner for the other three receptors (Graus-Porta et al., 1997; Yarden and Sliwkowski, 2001; Burgess et al., 2003; Garrett et al., 2003; Eccles, 2011; Roskoski, 2014; Wang, 2017). This heterodimerization is particularly important for ERBB2 signaling, as ERBB2 lacks a cognate ligand. Heterodimerization is also particularly important for ERBB3 signaling, as ERBB3 possesses markedly impaired kinase activity (Stern and Kamps, 1988; Wada et al., 1990; Carraway and Cantley, 1994; Olayioye et al., 2000; Bowers et al., 2001; Hynes et al., 2001; Yarden and Sliwkowski, 2001; Burgess et al., 2003; Earp et al., 2003; Zaczek et al., 2005; Citri and Yarden, 2006; Karamouzis et al., 2007; Mei and Xiong, 2008; Lemmon, 2009; Rudloff and Samuels, 2010; Eccles, 2011; Veikkolainen et al., 2011; Roskoski, 2014; Wang, 2017).
Receptor heterodimerization has profound functional implications; it is well established that the homodimer of a particular ERBB family member activates different signaling pathways and biologic effects than does a heterodimer containing (in part) the same particular ERBB family member. For example, heterodimerization of ERBB2 or EGFR with ERBB4 modifies the biologic response to ERBB4 ligands, illustrating that signaling by ERBB4 homodimers is different from signaling by ERBB4-EGFR or ERBB4-ERBB2 heterodimers (Beerli et al., 1995; Riese et al., 1995; Cohen et al., 1996; Karunagaran et al., 1996; Riese et al., 1996a; Zhang et al., 1996; Carraway et al., 1997; Chang et al., 1997; Graus-Porta et al., 1997; Weiss et al., 1997; Fitzpatrick et al., 1998; Pinkas-Kramarski et al., 1998b; Riese et al., 1998; Shelly et al., 1998; Wang et al., 1998; Hynes et al., 2001; Hobbs et al., 2002; Carpenter, 2003a; Earp et al., 2003; Hobbs et al., 2004; Zaczek et al., 2005; Muraoka-Cook et al., 2006; Mill et al., 2011a,b; Wilson et al., 2012b; Segers et al., 2020). Moreover, as discussed in detail in Section II.B and Section II.H, homotypic ERBB4 signaling by the 4ICD or by the constitutively active ERBB4 Q646C or I658E mutants is coupled to apoptosis, growth inhibition, and tumor suppression (Penington et al., 2002; Williams et al., 2003; Vidal et al., 2005; Gallo et al., 2006, 2013; Naresh et al., 2006; Pitfield et al., 2006; Vidal et al., 2007; Mill et al., 2011a; Arienti et al., 2019). In contrast, heterotypic signaling by ERBB4-EGFR and ERBB4-ERBB2 heterodimers is coupled to oncogenic phenotypes, including cell proliferation, migration, invasion, and chemoresistance (Cohen et al., 1996; Riese et al., 1996a; Zhang et al., 1996; Carraway et al., 1997; Olayioye et al., 1999, 2000; Mill et al., 2011b).
H. Trafficking of the ERBB4 Cytoplasmic Domain Enables Noncanonical Signaling in the Nucleus and the Mitochondria, Thereby Contributing to Apoptosis and Other Effects
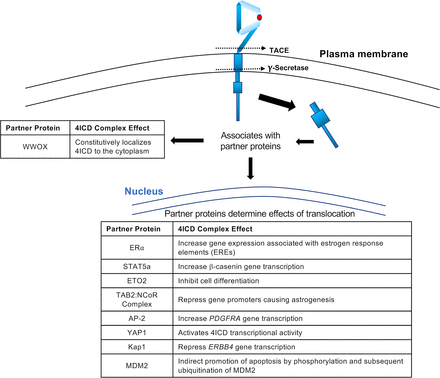
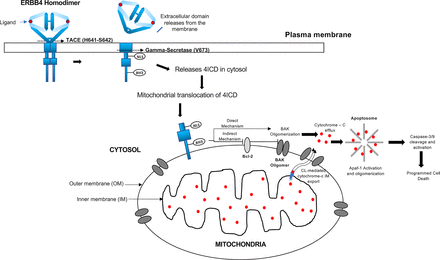
The binding of an ERBB4 full agonist (such as NRG1β or NRG2β) to ERBB4 can also trigger noncanonical signaling, which is commonly referred to as regulated intramembrane proteolysis (Fig. 5) (Carpenter, 2003a,b; Junttila et al., 2005; Citri and Yarden, 2006; Sardi et al., 2006; Schlessinger and Lemmon, 2006; Mei and Xiong, 2008; Paatero and Elenius, 2008; Blobel et al., 2009; Veikkolainen et al., 2011; Roskoski, 2014; Wang, 2017). In the first step, the transmembrane metalloprotease tumor necrosis factor (TNF) α-converting enzyme (ADAM17) cleaves the ERBB4 extracellular region near the transmembrane domain (between H641 and S642; Fig. 4), releasing the 120-kDa extracellular region into the extracellular milieu (Vecchi and Carpenter, 1997; Rio et al., 2000; Zhou and Carpenter, 2000; Ni et al., 2001; Carpenter, 2003a,b; Cheng et al., 2003; Schlessinger and Lemmon, 2006; Blobel et al., 2009). Parenthetically, this soluble form of the extracellular region of ERBB4 can function as a ligand sink (Gilmore and Riese, 2004).
Interactions of the 4ICD with partner proteins. The intracellular region of ERBB4 (4ICD) is depicted, along with the various partner proteins that are known to interact with the 4ICD in the nucleus and the cytoplasm. The biologic effect of each 4ICD complex is listed. The figure is not drawn to scale.
The remaining fragment of ERBB4, which consists of the transmembrane domain and the cytoplasmic domains, is cleaved by the gamma-secretase complex. The cleavage site appears to reside within the transmembrane domain, corresponding to the site of cleavage of Notch and the amyloid precursor protein by gamma-secretase (Fig. 4). This cleavage releases the 80-kDa cytoplasmic region of ERBB4 (the 4ICD) from the plasma membrane (Zhou and Carpenter, 2000; Ni et al., 2001, 2003; Lee et al., 2002a; Carpenter, 2003a,b; Linggi and Carpenter, 2006; Linggi et al., 2006; Naresh et al., 2006; Schlessinger and Lemmon, 2006; Greenwald and Kovall, 2013). This step requires the threonine-valine-valine (TVV) amino acid sequence (a PDZ recognition domain) present at the extreme carboxyl terminus of ERBB4 (Fig. 4) (Carpenter, 2003a; Ni et al., 2003; Blobel et al., 2009). This PDZ recognition domain may also mediate ERBB4 localization to basolateral membranes (Carraway and Sweeney, 2001). The PDZ recognition domain appears to be important to ERBB4 function; the addition of an epitope tag carboxyl-terminal to the TVV sequence disrupts the tumor suppressor activity of the ERBB4 Q646C mutant (Gallo et al., 2013).
The 4ICD induces growth arrest or apoptosis in a variety of tissues, including those of the breast (Zhu et al., 2006; Jones, 2008; Muraoka-Cook et al., 2008; Rokicki et al., 2010; Han et al., 2016; Arienti et al., 2019), brain (Sardi et al., 2006; Allison et al., 2011), and lung (Zscheppang et al., 2007; Liu et al., 2010; Hoeing et al., 2011). After release from the plasma membrane, the 4ICD traffics to the nucleus and alters patterns of gene expression that result in growth arrest (Srinivasan et al., 1999; Srinivasan et al., 2000; Ni et al., 2001; Lee et al., 2002a; Zhang et al., 2002; Carpenter, 2003b; Sardi et al., 2006; Blobel et al., 2009; Miller et al., 2017). Parenthetically, the trafficking of the 4ICD to the nucleus appears to be dependent on a putative nuclear localization sequence within the 4ICD (Fig. 4) (Ni et al., 2001; Lee et al., 2002a; Carpenter, 2003b; Williams et al., 2004; Gallo et al., 2013).
Nuclear 4ICD regulates gene transcription via various mechanisms (Ni et al., 2001; Carpenter, 2003b; Wang, 2017; Wang et al., 2019), including trafficking binding partners to the nucleus (Carpenter, 2003b; Long et al., 2003; Segers et al., 2020). This activity appears to be regulated in part by LXXLL motifs within the 4ICD (Fig. 4), which have been shown in other contexts to mediate interactions with nuclear hormone receptors (Heery et al., 1997; Edwards, 2000; Savkur and Burris, 2004) and one of which is required for the tumor suppressor activity of the ERBB4 Q646C mutant (Gallo et al., 2013). The functions of several transcriptional regulatory proteins are in some cases dependent on physical and functional interactions with the 4ICD, including the estrogen receptor (ER) α (Zhu et al., 2006; Blobel et al., 2009; Veikkolainen et al., 2011; Wang, 2017). Indeed, the 4ICD interacts with an ERα coactivator and increases the sensitivity of breast cancer cells to tamoxifen (Naresh et al., 2008; Wang et al., 2019). Additional transcriptional regulatory proteins whose functions are in some cases dependent on the 4ICD include the Yes-associated protein 1 (YAP1) (Komuro et al., 2003; Omerovic et al., 2004; Aqeilan et al., 2005; Citri and Yarden, 2006; Naresh et al., 2006; Blobel et al., 2009; Veikkolainen et al., 2011; Wang, 2017), STAT5a (Williams et al., 2004; Citri and Yarden, 2006; Blobel et al., 2009; Wang, 2017), eight twenty one 2 (ETO2) (Linggi and Carpenter, 2006; Blobel et al., 2009; Veikkolainen et al., 2011; Wang, 2017), the transforming growth factor beta activated kinase 1 binding protein 2-nuclear receptor corepressor (TAB2-NCoR) complex (Sardi et al., 2006; Schlessinger and Lemmon, 2006; Blobel et al., 2009; Veikkolainen et al., 2011; Wang, 2017), Krab-associated protein 1 (Gilmore-Hebert et al., 2010; Veikkolainen et al., 2011; Wang, 2017), and the activated enhancer-binding protein 2 (AP-2) (Sundvall et al., 2010; Veikkolainen et al., 2011; Wang, 2017) (Fig. 5).
WWOX binds to the proline-proline-alanine-tyrosine (PPAY) amino acid sequence found near the cytoplasmic carboxyl terminus of ERBB4 (Fig. 4), and increased ERBB4 tumor suppressor activity is associated with increased WWOX function (Aqeilan et al., 2007). Thus, it is not surprising that WWOX is also a tumor suppressor. WWOX appears to function in part by binding nuclear oncoproteins, including several transcription factors, and sequestering them in the cytoplasm (Aqeilan et al., 2005; Citri and Yarden, 2006; Blobel et al., 2009; Li et al., 2015; Pospiech et al., 2018); however, the tumor suppressor activity of WWOX may also be dependent on its 17b-hydroxysteroid dehydrogenase activity (Gluz et al., 2009; Krishnamurthy et al., 2012; Li et al., 2015; Pospiech et al., 2018). It has been reported that WWOX constitutively localizes the 4ICD to the cytoplasm (Aqeilan et al., 2007; Segers et al., 2020). However, this localization does not appear to solely account for the tumor suppressor activity of WWOX or ERBB4, as tumor suppression by ERBB4 seems to be dependent on 4ICD localization to the nucleus. Further complicating our understanding of the role that WWOX may play in mediating ERBB4 function, WWOX causes increased activity of the ataxia telangiectasia mutated (ATM) checkpoint kinase, leading to increased activity of the itchy E3 ubiquitin ligase (ITCH) and ubiquitination of Lys63 of WWOX. This results in increased nuclear translocation of WWOX and potentiation of further ATM activity in a positive feed-forward loop mechanism (Omerovic et al., 2007; Blobel et al., 2009; Schuchardt et al., 2013; Abu-Odeh et al., 2014; Aqeilan et al., 2014; Pospiech et al., 2018).
Our understanding of the functional consequences of ATM regulation by the 4ICD via WWOX and ITCH is impacted by the fact that the 4ICD causes G2/M cell cycle arrest and that this arrest is abrogated by 4ICD ubiquitination and degradation via interaction of the anaphase-promoting complex (APC) with a D-box sequence of the 4ICD (Fig. 4) (Strunk et al., 2007; Segers et al., 2020). This abrogation of G2/M arrest by APC-4ICD interactions may be related to the observation that ERBB4 induces G2/M arrest via a functional interaction with the breast cancer DNA repair associated 1 (BRCA1) protein (Muraoka-Cook et al., 2006; Segers et al., 2020). Moreover, our understanding of the role that WWOX plays in regulating ERBB4 function is also impacted by the fact that WWOX binding to ERBB4 inhibits YAP1 transcriptional activity (Aqeilan et al., 2005; Omerovic et al., 2007; Pospiech et al., 2018) as well as the fact that the 4ICD can also regulate the mouse double minute 2 (MDM2) E3 ubiquitin ligase (Arasada and Carpenter, 2005; Blobel et al., 2009), histone methylation, and human telomerase reverse transcriptase (Ishibashi et al., 2013; Segers et al., 2020). Finally, there are reports that the E3 ubiquitin ligases ITCH and WWP1 directly bind to and are specific for the ERBB4 Cyt1 isoforms and cause degradation of those isoforms. Furthermore, ITCH binds to the proline-proline-alanine-tyrosine-threonine-proline-methionine (PPAYTPM) amino acid sequence specific to Cyt1 (Fig. 4), suggesting that WWOX and ITCH compete for binding to ERBB4 (Omerovic et al., 2007; Sundvall et al., 2008; Li et al., 2009; Carraway, 2010).
The 4ICD may also trigger apoptosis by trafficking to the mitochondria and triggering the release of cytochrome C (Fig. 6) from the mitochondria, thereby stimulating programmed cell death. In some cases, ERBB4 expression and signaling are accompanied by mitochondrial accumulation of the 4ICD and binding of the proapoptotic Bcl-2 homologous antagonist/killer (BAK) protein to the 4ICD via the Bcl2 homology 3 (BH3)-like domain of the 4ICD (Fig. 4). This results in increased cytochrome C efflux (Naresh et al., 2006; Blobel et al., 2009; Miller et al., 2017; Arienti et al., 2019; Segers et al., 2020). This proposed mechanism is supported by the observation that when the ERBB4 V673I mutation abolishes ERBB4 cleavage by gamma-secretase, there is no accumulation of the 4ICD within the mitochondria, and the apoptotic activity associated with the 4ICD is abolished (Vidal et al., 2005).
The 4ICD regulates events in the mitochondria. The 4ICD translocates from the membrane to the outer mitochondrial membrane. Indirect and direct interactions of the 4ICD with BAK results in cytochrome C efflux and apoptotic cell death. The figure is not drawn to scale.
When the proapoptotic Bcl-2-associated X (BAX) or BAK proteins oligomerize and permeabilize the mitochondrial outer membrane, cytochrome C is exported from the inner mitochondrial membrane via the phospholipid cardiolipin, which is followed by efflux from the outer mitochondrial membrane (OMM) and into the cytosol via BAK oligomers. Cytochrome C complexes with the apoptosis protease-activating factor 1, causing its allosteric activation and oligomerization into an apoptosome. The activated cytochrome C cleaves nonmature caspase-3/9, which then triggers programmed cell death (Bossy-Wetzel et al., 1998; Zou et al., 1999; Garrido et al., 2006; Schug and Gottlieb, 2009). BH3-only proteins appear to stimulate mitochondrial-regulated apoptosis via indirect and direct mechanisms (Giam et al., 2008). In the indirect (or displacement) mechanism, BH3-only proteins bind Bcl-2 and related antiapoptotic proteins and inhibit their activity by sequestering them away from BAX or BAK, thereby allowing BAX or BAK oligomerization (Willis et al., 2007). The direct mechanism proposes that the BH3-only proteins directly interact with BAX or BAK, thereby stimulating their oligomerization and subsequent apoptotic signaling cascade (Kuwana et al., 2002; Letai et al., 2002).
The ERBB4 cytoplasmic region (4ICD) possesses a BH3-like domain (Fig. 4). This domain might allow the 4ICD to reside in the OMM, after which the BH3-like domain could either directly or indirectly stimulate apoptosis (Naresh et al., 2006; Wilfling et al., 2012). This apoptosis is dependent on BAK but not BAX (Naresh et al., 2006). One explanation is that BAK is found at higher levels in the OMM than is BAX. This may reflect the fact that BAX exists predominately in the cytosol but is translocated to the mitochondria after apoptotic stimulation (Naresh et al., 2006; Wilfling et al., 2012; Edlich, 2015). Therefore, the 4ICD could directly stimulate BAK oligomerization within the OMM. The 4ICD could also indirectly stimulate BAK oligomerization by displacing the antiapoptotic Bcl-XL, Bcl-w, myeloid leukemia cell differentiation (Mcl-1), or A1 proteins from BAK (Fig. 6).
The 4ICD may stimulate apoptosis via other mechanisms. For example, human MDM2 ubiquitinates TP53, thereby decreasing TP53 tumor suppressor activity and promoting cell survival. However, when 4ICD complexes with human MDM2, the complex is ubiquitinated and degraded, allowing for TP53 promotion of apoptotic pathways (Arasada and Carpenter, 2005; Veikkolainen et al., 2011). The array of direct and indirect tumor suppressor signaling pathways connected to 4ICD make elucidating the importance of the 4ICD difficult but critical to validating ERBB4 as a target for cancer therapeutics.
I. Transcriptional Splicing Isoforms of ERBB4 Confer Signaling Specificity
ERBB4 signaling specificity can be conferred by alternative splicing of the ERBB4 transcript. There are two sites of alternative splicing, resulting in four different isoforms (Fig. 4) (Elenius et al., 1997a; Carpenter, 2003a; Junttila et al., 2005; Sardi et al., 2006; Schlessinger and Lemmon, 2006; Chuu et al., 2008; Mei and Xiong, 2008; Paatero and Elenius, 2008; Veikkolainen et al., 2011; Segers et al., 2020). The first alternative splicing site affects the sequence of the extracellular juxtamembrane region, resulting in JMa and JMb isoforms (Gilbertson et al., 2001). The second alternative splicing site affects the cytoplasmic region carboxyl terminal (CT) to the kinase domain, resulting in the Cyt1 (CT-a) and Cyt2 (CT-b) isoforms (Kainulainen et al., 2000). Thus, ERBB4 can be transcribed into four isoforms: JMa-Cyt1, JMa-Cyt2, JMb-Cyt1, and JMb-Cyt2. The JMa-Cyt1 isoform is predominantly expressed and is considered the canonical ERBB4 transcript.
The ERBB4 transcriptional splicing isoforms possess distinct signaling activities. For example, the JMb isoforms lack the TNF converting enzyme cleavage site present in JMa isoforms. Hence, JMb isoforms cannot yield the 4ICD fragment whose intracellular trafficking is responsible for significant ERBB4 signaling activity (Rio et al., 2000; Sardi et al., 2006; Mei and Xiong, 2008; Paatero and Elenius, 2008; Segers et al., 2020) (see Section II.H for further details).
In contrast to the Cyt1 isoforms, the Cyt2 isoforms lack a short sequence of amino-acid residues from 1046 to 1061. This sequence includes a phosphorylation site (Tyr1056) that is part of a consensus sequence for binding the p85 regulatory subunit of PI3K (see Section II.F and Fig. 4 for further details). This sequence also contains a binding motif (PPAY) for proteins that contain a WW domain (see Section II.H and Fig. 4 for further details). Hence, as noted earlier, Cyt2 isoforms may be defective for coupling to PI3K signaling and WW proteins, including the WWOX tumor suppressor protein. These differences may be quantitative rather than absolute; WWOX binds to the Cyt2 isoform, albeit to a lesser extent than WWOX binding to the Cyt1 isoform (Kainulainen et al., 2000; Aqeilan et al., 2005; Mei and Xiong, 2008; Paatero and Elenius, 2008; Rudloff and Samuels, 2010; Schuchardt et al., 2013; Segers et al., 2020).
These and other differences in the signaling activities of the various ERBB4 isoforms also contribute to differences in biologic activities. For example, the JMa-Cyt2 isoform inhibits differentiation and promotes proliferation to a much greater extent than the JMb-Cyt2 isoform (Maatta et al., 2006; Sundvall et al., 2010; Veikkolainen et al., 2011). Furthermore, the region of ERBB4 that is specific to the Cyt1 isoforms (Fig. 4) and includes Tyr1056 is responsible for growth inhibition and differentiation of mammary ductal epithelial cells; this region is also required for the constitutively dimerized and active Q646C mutant of ERBB4 to function as a tumor suppressor (Muraoka-Cook et al., 2009; Wali et al., 2014; Segers et al., 2020). The Cyt1 isoform has recently been associated with an ERK-mediated negative-feedback mechanism that causes downregulation of ERBB4 activity (Haryuni et al., 2019).
However, the region of ERBB4 that is specific to Cyt1 isoforms and includes Tyr1056 is also required for the oncogenic activities of ERBB4, presumably through the action of ERBB4-EGFR or ERBB4-ERBB2 heterodimers (Junttila et al., 2005; Wali et al., 2014; Segers et al., 2020). Indeed, the 4ICD is associated with both tumor suppression and malignant phenotypes (see Section II.H for further details) (Junttila et al., 2005; Fujiwara et al., 2014); these functions suggest that ERBB4 homodimerization and heterodimerization cause differential phosphorylation of the ERBB4 cytoplasmic domain and differential association with effector proteins, resulting in distinct biologic responses.
J. Summary and Implications for ERBB4 Function in Human Malignancies
It appears that ERBB4 homodimers function as tumor suppressors, whereas ERBB4-EGFR and ERBB4-ERBB2 heterodimers function as oncoproteins. However, these do not appear to be absolute relationships, as the function of ERBB4 homodimers and heterodimers can be influenced by the involvement of ERBB4 splicing isoforms and of ERBB4 ligands, each of which can confer signaling and biologic specificity. Nonetheless, in contexts in which ERBB4 heterodimers predominate over ERBB4 homodimers, one would expect that increases in ERBB4 or ligand expression and ERBB4 gain-of-function mutants would be associated with malignancies or more aggressive tumor phenotypes. Conversely, in contexts in which ERBB4 homodimers predominate over ERBB4 heterodimers, one would expect that decreases in ERBB4 or ligand expression and ERBB4 loss-of-function mutants would be associated with malignancies or more aggressive tumor phenotypes. Below we will describe how ERBB4 expression, ligand expression, and ERBB4 mutations align with this mechanistic model for ERBB4 function in different types of tumors.
III. Tumors in Which ERBB4 Appears to Function as a Tumor Suppressor
A. Introduction
In the following subsections, we will discuss the different types of tumors in which ERBB4 appears to function as a tumor suppressor. In these types of tumors, ERBB4 expression is typically low. Likewise, only a small fraction of cell lines established from these types of tumors exhibits detectable ERBB4 transcription. In many instances, a decrease in ERBB4 copy number underlies the low level of ERBB4 expression (Segers et al., 2020). Loss-of-function mutations in ERBB4 are a potential mechanism for disrupting ERBB4 tumor suppressor activity. However, loss-of-function mutations in ERBB4 have yet to be validated as bona fide tumor drivers.
B. Bladder
In general, ERBB4 expression is lower in transitional cell carcinomas than in normal urothelium (Rotterud et al., 2007). Low ERBB4 expression is associated with high-grade tumors and shorter survival (Black and Dinney, 2008; Kassouf et al., 2008; Segers et al., 2020). Likewise, elevated ERBB4 expression in urothelial tumors is associated with lower-grade tumors, less invasive tumors, and a more favorable prognosis (Memon et al., 2004; Memon et al., 2006; Black and Dinney, 2008; Segers et al., 2020). These data suggest that ERBB4 functions as a tumor suppressor in these tumors. Superficially, this hypothesis appears to be at odds with the observation that EREG expression is elevated in bladder cancer samples and that EREG overexpression is associated with increased metastatic potential (Thogersen et al., 2001; Nicholson et al., 2004; Riese and Cullum, 2014). However, given that EREG is a high-affinity ligand for both EGFR and ERBB4, it is possible that the oncogenic activity of EREG is mediated by EGFR rather than by ERBB4.
C. Liver
ERBB4 is expressed in 39%–63% of cholangiocarcinomas (tumors of the biliary tract), and this expression is associated with a more favorable prognosis in intrahepatic cholangiocarcinomas that lack EGFR expression (Ito et al., 2001b; Yang et al., 2014; Pellat et al., 2018; Segers et al., 2020). The loss of ERBB4 expression may play a role in the progression of benign hepatic lesions to hepatocellular carcinomas (HCCs) (Lee et al., 2007; Segers et al., 2020). This hypothesis is consistent with the observation that ERBB4-null hepatocytes exhibit a higher rate of proliferation than control hepatocytes (Liu et al., 2017; Segers et al., 2020) and is consistent with the observation that ERBB4 expression is frequently lower in HCCs than in the normal hepatocytes. There are contradictory results regarding the effect of ERBB4 expression on HCC outcome (Ito et al., 2001a; Uberall et al., 2008; Liu et al., 2017; Segers et al., 2020). Nonetheless, taken together these data indicate that ERBB4 is functioning as a tumor suppressor in intrahepatic cholangiocarcinomas and HCCs.
D. Prostate
ERBB4 is strongly expressed by normal prostate luminal cells. In contrast, ERBB4 protein expression is detected in only 23% of prostate cancer specimens, and no human prostate cell lines tested to date exhibit detectable ERBB4 protein expression (Grasso et al., 1997; Uberall et al., 2008). Furthermore, the constitutively active ERBB4 Q646C and I658E mutants function as tumor suppressors in human prostate cell lines (Williams et al., 2003; Vidal et al., 2007). Superficially, these results are at odds with the observation that exogenous ERBB4 expression in androgen-independent human prostate tumor cell lines causes resistance to tyrosine kinase inhibitors (Carrion-Salip et al., 2012). However, as noted elsewhere, heterodimerization of ERBB4 with EGFR or ERBB2 (as a result of chemotherapy-induced EGFR or ERBB2 overexpression) may enable ERBB4 to cause this chemoresistance.
IV. Tumors in Which ERBB4 Appears to Function as an Oncoprotein
A. Introduction
There are multiple reports in which an ERBB4 ligand stimulates malignant phenotypes and chemoresistance of tumor cells (Yamano et al., 2010; Mill et al., 2011a; Sato et al., 2013; Dahlhoff et al., 2014; Riese and Cullum, 2014; Mota et al., 2017; Segers et al., 2020). However, many ERBB4 ligands also bind either EGFR or ERBB3. Moreover, ligand binding to ERBB4 can induce ERBB4 homodimerization or ERBB4 heterodimerization with other ERBB receptors. Thus, reports that an ERBB4 ligand stimulates malignant phenotypes and chemoresistance of tumor cell lines should not be viewed as strictly contradictory of the evidence that ERBB4 functions as a tumor suppressor. Indeed, in the following subsections, we will provide evidence that ERBB4-ERBB2 and ERBB4-EGFR heterodimers function as oncoproteins. Oncogenic activity of these heterodimers may result from overexpression of ERBB4, ERBB2, or EGFR or gain-of-function mutations in ERBB4, ERBB2, or EGFR.
B. Brain
ERBB4 overexpression is associated with a statistically insignificant decrease in overall survival among patients with childhood medulloblastoma. However, the combination of ERBB2 and ERBB4 overexpression is associated with a dramatic reduction in overall survival that is greater than the effects of ERBB2 alone (Gilbertson et al., 1997; Gilbertson et al., 1998; Gilbertson et al., 2001; Carpenter, 2003a; Rickert, 2004; Rickert and Paulus, 2005; Britsch, 2007). NRG1β and ERBB4 are expressed in the developing cerebellum, but ERBB2 is not. Taken together, these data suggest that ERBB2 overexpression acquired during tumorigenesis or tumor progression enables the pre-existing ERBB4 and NRG1β expression to drive signaling by oncogenic ERBB4-ERBB2 heterodimers and the development of medulloblastomas in the cerebellum (Gilbertson et al., 1998; Britsch, 2007).
Molecular profiling has enabled the assignment of medulloblastomas into four main subtypes: Wingless, Sonic Hedgehog, group 3, and group 4 (Northcott et al., 2012; Rahmann and Gilbertson, 2018). An integrative proteogenomic approach reveals that group 4 tumors commonly exhibit elevated expression of ERBB4 and its ligand NRG2 as well as hallmarks of elevated receptor tyrosine kinase signaling, including ERK and PI3K signaling. Surprisingly, this report did not indicate that ERBB2 expression was elevated in the group 4 tumors, raising questions about the mechanism by which ERBB4 may be coupled to tumorigenesis or tumor progression in group 4 medulloblastomas (Forget et al., 2018; Rahmann and Gilbertson, 2018).
ERBB2 and ERBB4 are coexpressed in most childhood ependymomas, and high levels of coexpression are associated with elevated tumor proliferation. Likewise, limited data suggest that elevated ERBB2 and ERBB4 coexpression is associated with less favorable clinical outcomes (Gilbertson et al., 2002; Carpenter, 2003a).
Limited evidence exists regarding the role of ERBB4 in glioblastomas. Elevated ERBB4 phosphorylation is associated with shorter survival in patients with glioblastoma (Donoghue et al., 2018; Segers et al., 2020). In contrast, ERBB4 copy number is frequently reduced across glioblastoma cell lines (Jones et al., 2018; Segers et al., 2020). One possible explanation for these apparently contradictory results is that ERBB4 heterodimers may function as oncoproteins in glioblastoma, whereas ERBB4 homodimers may function as tumor suppressors in glioblastoma. There are reports that ERBB4 does not play a major role in gliomas (Berezowska and Schlegel, 2011). Moreover, ERBB4 expression is higher in low-grade (less aggressive) gliomas than in high-grade (more aggressive) gliomas. Thus, there is little evidence that ERBB4 functions as an oncoprotein in gliomas. In contrast, ERBB4 overexpression has been observed in meningiomas. This result suggests that ERBB4 is an oncoprotein in meningiomas (Andersson et al., 2004; Uberall et al., 2008).
C. Colon
EGFR is a well established colorectal cancer (CRC) biomarker and target for therapeutic intervention. In contrast, there is considerably less information about the role that ERBB4 plays in colorectal cancer (Khelwatty et al., 2013; Mitsui et al., 2014; Williams et al., 2015; Segers et al., 2020). Nonetheless, ERBB4 overexpression or elevated phosphorylation is observed in all stages of CRC and has been reported to be associated with more aggressive colorectal tumors, particularly metastatic behavior (Kountourakis et al., 2006; Baiocchi et al., 2009; Frey et al., 2010; Khelwatty et al., 2013; Mitsui et al., 2014; Williams et al., 2015; Mota et al., 2017; Segers et al., 2020). ERBB4 copy number does not appear to be altered in a significant fraction of CRC samples; therefore, overexpression of ERBB4 seems to reflect changes in transcription or protein stability (Segers et al., 2020).
Nonsynonymous ERBB4 mutations are found in 7.5%–11% of CRCs, and ∼1.5% of CRCs are predicted to harbor an ERBB4 tumor driver mutation (Mishra et al., 2017; Segers et al., 2020). Nonetheless, only a few ERBB4 mutations in CRC samples have been described (V721I, P854Q, D861Y, I1030M; Tables 1 and 2). Of these, the D861Y mutant exhibits reduced agonist-dependent and -independent tyrosine phosphorylation in a heterologous model system (Soung et al., 2006). On the surface, this finding is inconsistent with the hypothesis that ERBB4 functions as a tumor suppressor. However, given that these experiments were performed in the context of the noncanonical JMa-Cyt2 isoform, the relevance of this finding is unclear (Tvorogov et al., 2009).
ERBB4 missense mutations that are found in human tumor samples and whose function has been at least partially characterized.
ERBB4 signaling protects colon epithelial cells from TNF-induced apoptosis, and reduced ERBB4 expression is associated with decreased cell proliferation and increased apoptosis (Frey et al., 2009; Segers et al., 2020). Similarly, knockdown of ERBB4 expression in a CRC cell line inhibits anchorage-independent proliferation (Williams et al., 2015; Segers et al., 2020). Likewise, ectopic ERBB4 expression is associated with the development of resistance to EGFR inhibitors, such as cetuximab (Bae et al., 2014; Sun et al., 2017).
ERBB4 heterodimers may be responsible for the oncogenic activity of ERBB4, as EGFR is frequently expressed in colorectal tumors. Furthermore, ERBB4 is found in the membrane of CRC cells, which is suggestive of heterotypic signaling (Mitsui et al., 2014). In contrast, ERBB4 localization in the cytoplasm or nucleus is suggestive of homotypic signaling. Consistent with this model for ERBB4 function, coexpression of ERBB4 and ERBB2 (Lee et al., 2002b; Khelwatty et al., 2013) and coexpression of ERBB4 and ERBB3 are each associated with late-stage colorectal tumors (Ljuslinder et al., 2009; Khelwatty et al., 2013). A potential mechanism for the coexpression of ERBB3 and ERBB4 is revealed by ApcMin transgenic mice in which ERBB3 is ablated in the intestine. Such animals exhibit a loss of ERBB4 expression and a dramatic reduction of intestinal tumors. Moreover, in a human colon cancer cell line containing a gain-of-function mutant Kirsten RAS (KRAS) allele, reduced ERBB3 expression is associated with decreased ERBB4 expression and small interfering RNA against either ERBB3 or ERBB4 results in increased apoptosis. Thus, either ERBB4-EGFR or ERBB4-ERBB4 heterodimers have been postulated to be responsible for the oncogenic activity of ERBB4 in colorectal tumors (Lee et al., 2009). The functional difference between ERBB4 homodimers and ERBB4 heterodimers may account for the observation that ERBB4 attenuates inflammation in colitis and may therefore function as a tumor suppressor in colitis-associated CRC (Schumacher et al., 2017; Segers et al., 2020). On the other hand, given that the promotion of colon adenoma to carcinoma by an intestine-specific KA11 C-terminal interacting tetraspanin (KITENIN) transgene (and ApcMin) is accompanied by elevated expression of the noncanonical ERBB4 CT-b isoforms, it is possible that differences in ERBB4 function observed in the colonic epithelium are isoform-specific (Bae et al., 2016).
D. Stomach
ERBB4 is frequently amplified in gastric cancer and is associated with advanced stage (He et al., 2015; Arienti et al., 2019) and poorer prognosis (Shi et al., 2012; Qu et al., 2013; Segers et al., 2020). The microRNA (miR) miR-551b inhibits ERBB4 gene expression. Thus, low levels of miR-551b expression are associated with a poorer prognosis in gastric cancer patients (Song et al., 2017). Likewise, the irreversible pan-ERBB tyrosine kinase inhibitor dacomitinib yields some disease control in patients whose gastric tumors overexpress ERBB2, which is consistent with the hypothesis that ERBB4-ERBB2 heterodimers function as oncoproteins (Desai et al., 2013; Mishra et al., 2017).
There is limited evidence that ERBB4 mutations may contribute to stomach cancer. The ERBB4 N548T mutation (Zang et al., 2012; Qu et al., 2013) and A773S mutation (Soung et al., 2006; Mishra et al., 2017) have each been found in a single gastric cancer sample. In a cohort of 294 stomach adenocarcinoma samples derived from northern Chinese patients, 20 samples contained one or more ERBB4 mutations (Table 2). One of these ERBB4 mutations alters an amino-acid codon (Arg106) that is also mutated in melanoma cases in The Cancer Genome Atlas Skin Cutaneous Melanoma (TCGA-SKCM) dataset and in stomach adenocarcinoma cases in the TCGA Stomach Adenocarcinoma (STAD) dataset (Chen et al., 2015). Altogether, the northern Chinese stomach adenocarcinoma datasets and the TCGA-STAD dataset contained 54 distinct ERBB4 missense mutations (Table 2). The same cohort of 294 northern Chinese gastric adenocarcinomas contains numerous samples that contain NRG1 mutations. Moreover, an NRG1 mutation is less common in samples that contain an ERBB4 mutation and vice versa (Chen et al., 2015). Taken together, these data suggest that increased ERBB4 signaling as a result of gain-of-function tumor driver mutations in the NRG1 or ERBB4 genes may contribute to gastric cancer genesis and/or progression.
ERBB4 missense mutations that are found in human tumor samples, yet whose function is largely unknown.
E. Head and Neck
ERBB4 is overexpressed in a significant fraction of head and neck squamous cell carcinomas (HNSCCs). This overexpression is observed in both in situ and invasive tumors (Kalyankrishna and Grandis, 2006; Uberall et al., 2008). The overexpression of EGFR and its ligands has been well documented in head and neck cancers, but overexpression of EGFR ligands is more effective in predicting disease prognosis. For example, AREG, EGF, HBEGF, and BTC overexpression are each independently associated with reduced 5-year survival, although AREG overexpression is correlated with HBEGF overexpression (Kalyankrishna and Grandis, 2006; Uberall et al., 2008; Ahsan et al., 2010; Gao et al., 2016).
HNSCCs devoid of human papillomaviruses (HPVs) exhibit greater AREG expression than HPV-positive tumors (Gao et al., 2016). Similarly, HPV-negative tumors exhibit greater EREG expression than do HPV-positive tumors. HPV infection is associated with methylation of the EREG promoter and reduced EREG transcription; reversing this methylation causes increased EREG transcription (Khanal et al., 2020). Taken together, these data suggest that HPVs and EGF family members independently drive the genesis and progression of HNSCCs. Recall that BTC, EREG, and HBEGF are ligands for both EGFR and ERBB4. Moreover, ligand stimulation can cause EGFR heterodimerization with ERBB4. Therefore, it is plausible to postulate that ERBB4 potentiates EGFR function during the genesis and progression of HNSCCs.
F. Lung
ERBB4 expression is detected in several human lung cancer cell lines, and silencing ERBB4 decreases the viability of these cell lines. Ibrutinib, a broad-specificity tyrosine kinase inhibitor that has high affinity for the Bruton’s tyrosine kinase also inhibits ERBB4, albeit with much less potency. Ibrutinib inhibits ERBB4 tyrosine phosphorylation in several human lung cancer cell lines and modestly inhibits proliferation by the H661 human lung cancer cell line in mouse xenografts. It is reasonable to postulate that the modest effect of ibrutinib on H661 cells in vivo may reflect the relatively low affinity of ibrutinib for ERBB4 (Rauf et al., 2018).
The Y285C, D595V, D931Y, and K935I ERBB4 mutants found in non–small cell lung carcinoma samples (Table 1) exhibit increased ligand-dependent and -independent tyrosine phosphorylation and increased heterodimerization with ERBB2. Furthermore, the Y285C, D595V, and K935I mutants caused increased survival of NIH 3T3 cells in the absence of serum. The ERBB4 G802dup mutation has been found in large-cell lung carcinomas (Table 1). The relevance of the G802dup mutation to lung cancer is unclear, particularly since it causes decreased ligand-dependent ERBB4 tyrosine phosphorylation. Additional ERBB4 missense mutations (N181S, T244R, R306S, V348L, H618P, R782Q, E810K, T926M; Table 2) have been identified in various forms of lung cancer, but it remains unclear whether these mutations are functionally significant (Soung et al., 2006; Ding et al., 2008; Tvorogov et al., 2009; Kurppa et al., 2016; Mishra et al., 2017; Segers et al., 2020).
Finally, overexpression of the EGFR and ERBB4 ligand EREG is frequently observed in lung adenocarcinomas and is associated with shorter disease-free survival and overall survival. For example, patients with lung adenocarcinoma with elevated EREG expression and an activating mutation in KRAS exhibit a poorer prognosis than patients who possessed only one of these factors (Sunaga et al., 2013; Sunaga and Kaira, 2015). If indeed EREG and an activated KRAS allele cooperate to drive more aggressive forms of lung adenocarcinoma, it is reasonable to postulate that EREG stimulation of EGFR coupling to RAS signaling is not sufficient for these adenocarcinomas and that EREG stimulation of ERBB4 signaling (via an ERBB4-EGFR heterodimer) is required for these aggressive tumors.
G. Bone
ERBB4 is constitutively phosphorylated in early passage human osteosarcoma cell lines. The 4ICD is observed in the nuclei of osteosarcoma clinical samples and cell lines, which is surprising given that the 4ICD is indicative of homotypic ERBB4 signaling and tumor suppressor activity. Furthermore, ERBB4 knockdown inhibits the proliferation and anchorage independence of osteosarcoma cell lines and increases the sensitivity of osteosarcoma cell lines to cytotoxic chemotherapeutic agents (Hughes et al., 2004, 2006; Wang et al., 2018, 2019).
H. Ovary
ERBB4 transcription and ERBB4 protein expression are higher in ovarian cancer samples than in borderline ovarian tumors or benign ovarian tumors. ERBB2 transcription and ERBB2 protein expression are likewise elevated but not EGFR transcription or EGFR protein expression (Steffensen et al., 2008; Wang, 2017). Taken together, these data suggest that ERBB4-ERBB2 heterodimers contribute to ovarian malignancies rather than ERBB4 homodimers or ERBB4-EGFR heterodimers. Overexpression of the ERBB4 JMa-Cyt1 isoform is associated with increased grade and poorer overall survival in patients with ovarian cancer (Paatero et al., 2013; Wang et al., 2019). This increase in endogenous ERBB4 expression appears to result from an increase in ERBB4 copy number in ovarian tumor samples (Segers et al., 2020). The increase in ERBB4 copy number and consequent increase in ERBB4 JMa-Cyt1 transcription appear to be functionally significant; ectopic overexpression of the ERBB4 JMa-Cyt1 isoform increases the anchorage-independent growth of ovarian cancer cell lines, but the JMa-Cyt2 isoform does not (Paatero et al., 2013; Wang et al., 2019).
I. Thyroid
ERBB4, HBEGF, and NRG1 expression are increased in some thyroid cancers relative to non-neoplastic thyroid tissue. Immunohistochemical detection of the ERBB4 protein reveals increased expression of ERBB4 in both the cytoplasm and membrane in papillary thyroid carcinomas, suggesting that the ERBB4 protein is functional and mediates HBEGF and NRG action. Interestingly, there is some evidence that ERBB4 overexpression is inversely correlated with the V600E hotspot mutation in the v-Raf murine sarcoma viral oncogene homolog B (BRAF) gene, suggesting that ERBB4 and BRAF may independently drive tumor progression (Haugen et al., 1996; Fluge et al., 2000; Kato et al., 2004; Wiseman et al., 2008; Ota et al., 2013; Schulten et al., 2015; Zhang et al., 2018).
HBEGF and ERBB4 are overexpressed in malignant thyroid tissues relative to both benign thyroid tumors and normal thyroid tissue. In contrast, EGFR is overexpressed in both malignant and benign thyroid tumors. In vitro, HBEGF stimulates the motility of thyroid cancer cell lines. Treatment with the ERBB4 and EGFR tyrosine kinase inhibitor N-(3-chlorophenyl)-6,7-dimethoxy-4-quinazolinamine attenuates this activity. Taken together, these data suggest that ERBB4 may heterodimerize with EGFR to mediate HBEGF-induced metastatic activity in thyroid cancers (Ota et al., 2013). Elevated ERBB4 expression in some papillary thyroid carcinoma cells appears to be the consequence of miR-326 downregulation. Indeed, ectopic expression of miR-326 in papillary thyroid carcinoma cell lines suppresses tumorigenesis in vivo. However, care must be taken not to overinterpret this result, as miR-326 also inhibits MAPK expression (Nie et al., 2020).
Finally, in a cohort of poorly differentiated thyroid carcinomas, nine different ERBB4 missense mutations were found (R106S, D113V, D165N, I166N, C213Y, G219D, E233G, C589Y, and C614Y; Table 2) (Gerber et al., 2018). The functional significance of these ERBB4 mutants is not known; nonetheless, their existence is consistent with the hypothesis that ERBB4 functions as an oncogene in thyroid cancers.
J. Hematopoietic
ERBB4 is not commonly believed to be expressed in normal cells of the hematopoietic lineage. ERBB4 knockout mice experience midembryonic lethality due to failed development of myocardial trabeculae (Gassmann et al., 1995). ERBB4 knockout mice in which ERBB4 expression in the heart is restored are viable at birth and exhibit normal cardiac development. However, these animals exhibit defects in mammary gland maturation, cranial neural crest migration, cranial nerve architecture, and cerebellum defects. Yet, there are no reports that these animals exhibit deficits in hematopoiesis or hematopoietic cell function (Tidcombe et al., 2003).
Thus, it is surprising that ERBB4 expression is observed during human erythroid cell maturation and that the irreversible ERBB tyrosine kinase inhibitor neratinib inhibits erythroid maturation in mice. Likewise, morpholino inhibition of ERBB4 in zebrafish embryos decreases embryonic erythropoiesis (Kinney et al., 2019a,b). Moreover, elevated expression of ERBB4 is detected in 24% of anaplastic large-cell lymphomas (ALCLs) that lack a chromosomal translocation that affects the anaplastic lymphoma kinase gene. The ERBB4 protein expressed in these samples is truncated as a result of cryptic transcriptional promotion from within an intron in the ERBB4 gene. Most ALCLs that exhibit expression of the truncated ERBB4 exhibit a morphology reminiscent of Hodgkin lymphoma; this morphology is typically rare in ALCLs. Moreover, patients that are ERBB4-positive with ALCL have overall survival rates inferior to those with the anaplastic lymphoma kinase translocation (Gaulard and de Leval, 2016; Scarfo et al., 2016).
The truncated ERBB4 protein, which consists of the carboxyl-terminal cytoplasmic domain and has an apparent molecular weight of 50 kDa, is constitutively tyrosine-phosphorylated. The truncated ERBB4 protein can transform the growth of NIH-3T3 fibroblasts. Patient-derived tumor cells that endogenously express the truncated ERBB4 protein were established. Neratinib kills these patient-derived tumor cells in ex vivo and in vivo settings (Gaulard and de Leval, 2016; Scarfo et al., 2016). Overall, these data suggest that ERBB4 is an oncogene in a subset of ALCLs, although it is not apparent whether ERBB4 functions autonomously or through heterodimerization with another ERBB receptor.
V. Tumors in Which ERBB4 Appears to Function as an Oncoprotein and as a Tumor Suppressor
A. Pancreas
Elevated ERBB4 expression is associated with favorable tumor staging (Thybusch-Bernhardt et al., 2001; Segers et al., 2020). Similarly, relative to normal pancreatic tissue, malignant pancreatic tissue frequently exhibits reduced ERBB4 expression (te Velde et al., 2009; Mill et al., 2011a). Expression of the constitutively homodimerized and constitutively active ERBB4 Q646C mutant inhibits clonogenic proliferation in pancreatic tumor cell lines that lack endogenous ERBB4 expression (Mill et al., 2011a). Taken together, these findings suggest that ERBB4 functions as a pancreatic tumor suppressor gene.
In contrast, the expression of wild-type ERBB4 in the pancreatic tumor cell lines that lack endogenous ERBB4 expression enables the ERBB4 agonist NRG1β to stimulate anchorage-independent growth (Mill et al., 2011a). Likewise, the ERBB4 ligand BTC is overexpressed in pancreatic tumor cell lines and stimulates the proliferation of pancreatic tumor cell lines (Yokoyama et al., 1995; Mashima et al., 1996; Dunbar and Goddard, 2000; Kawaguchi et al., 2000; Li et al., 2003; Dahlhoff et al., 2014).
Pancreatic fibrosis is a major component of several diseases of the pancreas, including pancreatic cancer. In a transgenic mouse, pancreatic-specific expression of Hbegf causes pancreatic fibrosis. This effect is significantly reduced in a bitransgenic mouse that possesses an EGFR mutant that exhibits a partial loss of tyrosine kinase activity (Blaine et al., 2009). However, this result may not mean that Hbegf functions solely through EGFR, as disrupting EGFR kinase activity causes reduced proliferative signaling by ERBB4-EGFR heterodimers (Wilson et al., 2012b).
In a genetically engineered mouse, pancreas-specific knockout of BTC in the context of a KRAS G12D mutant results in decelerated progression of pancreatic ductal adenocarcinoma (PDAC). Similarly, pancreas-specific overexpression of BTC in the context of a KRAS G12D mutant results in accelerated progression of PDAC. Knockout of EGFR, ERBB2, or ERBB4 diminishes this acceleration (Hedegger et al., 2020). These results suggest that ERBB4 functions as a PDAC oncoprotein in the context of a KRAS activating mutation through heterodimerization with another ERBB receptor (Mill et al., 2011a).
B. Breast
ERBB4 ligands inhibit proliferation and promote differentiation in several human breast cancer cell lines (Sartor et al., 2001; Carpenter, 2003a; Muraoka-Cook et al., 2006; Karamouzis et al., 2007; Uberall et al., 2008; Arienti et al., 2019; Segers et al., 2020). These data are consistent with the observation that ERBB4 signaling contributes to mammary gland differentiation rather than expansion (Schroeder and Lee, 1998; Jones et al., 1999; Carpenter, 2003a; Long et al., 2003; Tidcombe et al., 2003; Eccles, 2011; Segers et al., 2020). This inhibition of proliferation and contribution to differentiation appear to be specific for the ERBB4 isoform; in transgenic mice, breast-specific expression of the canonical CT-a isoform of the 4ICD causes decreased proliferation of the ductal epithelium and lactogenic differentiation, whereas breast-specific expression of the CT-b isoform of the 4ICD causes epithelial hyperplasia (Muraoka-Cook et al., 2009).
Reduced ERBB4 expression is predictive of recurrence of ductal carcinoma in situ (Barnes et al., 2005; Uberall et al., 2008; Lyu et al., 2018) and reduced overall survival among patients with breast cancer (Das et al., 2010; Wang et al., 2019). Similarly, ERBB4 overexpression is associated with improved relapse-free survival (Pawlowski et al., 2000; Wang et al., 2016; Segers et al., 2020), disease-free survival (Koutras et al., 2008; Sassen et al., 2008), and overall survival (Pawlowski et al., 2000; Witton et al., 2003; Junttila et al., 2005; Koutras et al., 2008; Sassen et al., 2008). ERBB4 expression is positively correlated with estrogen receptor (ER) and progesterone receptor (PR) expression, raising some concern about ERBB4 status being an independent predictor of breast cancer outcome (Bacus et al., 1996; Knowlden et al., 1998; Pawlowski et al., 2000). Indeed, ERBB4 overexpression more strongly correlates with a favorable outcome in ER/PR-positive breast cancer cases than in ER/PR-negative breast cancer cases (Junttila et al., 2005). One potential explanation for this observation is that homotypic ERBB4 signaling by the 4ICD mediates tamoxifen-induced apoptosis (Naresh et al., 2008). The expression of ERBB4 and the tumor suppressor WWOX are both significantly lower in breast tumors derived from lymph node metastases than in the matched primary tumors (Aqeilan et al., 2007; Pospiech et al., 2018). It has been proposed that the loss of ERBB4 expression observed in malignant samples is due to ERBB4 promoter hypermethylation, as treatment of the BT20 human breast tumor cell line with a DNA demethylating agent causes decreased methylation of the ERBB4 promoter, increased ERBB4 expression, and increased apoptosis (Das et al., 2010; Segers et al., 2020). Taken together, these data suggest that ERBB4 functions as a breast cancer tumor suppressor gene, particularly in ER/PR-positive/HER2-negative breast cancers (Bacus et al., 1996; Vogt et al., 1998; Suo et al., 2002; Witton et al., 2003; Zaczek et al., 2005; Muraoka-Cook et al., 2008; Sundvall et al., 2008; Koutras et al., 2010; Segers et al., 2020).
In contrast, there are numerous reports that ERBB4 ligands stimulate proliferation, malignant phenotypes, and chemoresistance in models of breast cancer (Karamouzis et al., 2007; Eccles, 2011; Xia et al., 2013; Feldinger and Kong, 2015). Similarly, BTC expression in breast tumor samples is associated with a poor clinical outcome (Olsen et al., 2012; Dahlhoff et al., 2014). ERBB4 underexpression is associated with improved responsiveness to endocrine therapies and longer relapse-free survival (Bieche et al., 2003; Wege et al., 2018; Brockhoff, 2019). Likewise, silencing endogenous ERBB4 expression in the ZR-75-1 human breast tumor cell line causes increased sensitivity to tamoxifen (Wege et al., 2018). Parenthetically, we postulate that this sensitivity is due to decreased signaling by ERBB4 heterodimers, as increased homotypic ERBB4 signaling also causes increased sensitivity to tamoxifen (Naresh et al., 2008). Another finding that indicates that increased ERBB4 signaling is coupled to tumorigenesis or tumor progression is that ERBB4 overexpression is associated with chemoresistance (including resistance to endocrine agents) and poorer prognosis (Lodge et al., 2003; Sutherland, 2011; Kim et al., 2016; Wege et al., 2018; Segers et al., 2020). Moreover, ectopic ERBB4 overexpression can stimulate proliferation and anchorage independence (Junttila et al., 2005).
The proliferative effects of ERBB4 have been observed in transgenic model systems. For example, in mice that exhibit high levels of canonical ERBB4 expression in the breast due to introduction of the MMTV-ERBB4 (JMa/CT-a) transgene, mammary terminal end bud differentiation is suppressed, and neoplastic mammary lesions form (Wali et al., 2014). These effects contrast those of the 4ICD CT-a transgene, which, as discussed earlier, causes terminal differentiation and suppresses proliferation (Muraoka-Cook et al., 2009). The functional difference between the full-length ERBB4 JMa/CT-a transgene and the 4ICD CT-a transgene appears to be due to the fact that full-length ERBB4 can heterodimerize with another ERBB family receptor, whereas the 4ICD participates only in homotypic ERBB4 signaling.
It is well accepted that many triple-negative breast tumors (particularly those of the basal-like subtype of triple-negative breast cancer) exhibit elevated expression of EGFR and its ligands (Farnie et al., 2007; Kenny and Bissell, 2007; Gluz et al., 2009; Viale et al., 2009; Burness et al., 2010; Foley et al., 2010). Triple-negative breast cancers (TNBCs) exhibit significantly increased ADAM17 expression, which may stimulate ERBB4 signaling by enhancing the release of NRGs and BTC into the extracellular milieu. In TNBCs, ERBB4 overexpression is associated with a poorer prognosis, thereby supporting our hypothesis that ERBB4-EGFR heterodimers function as oncoproteins in TNBCs (McGowan et al., 2013; Feldinger and Kong, 2015; Kim et al., 2016; Wang et al., 2019).
ERBB2 is a well established breast cancer oncogene. In transgenic mice that possess the MMTV-ERBB2 transgene, breast tumorigenesis occurs with relatively slow kinetics and is accompanied by overexpression of endogenous EGFR, ERBB3, and ERBB4 and phosphorylation of protein kinase B (PKB or AKT1). In contrast, in transgenic mice that possess the MMTV-ERBB2 transgene and a constitutively active AKT1 allele under control of the MMTV promoter (MMTV-caAKT1), tumors form with rapid kinetics and exhibit elevated AKT1 phosphorylation but not overexpression of endogenous EGFR, ERBB3, and ERBB4. Thus, it appears that tumorigenesis driven by ERBB2 requires AKT1 phosphorylation, which occurs through overexpression of EGFR, ERBB3, and ERBB4 and heterodimerization of these receptors with ERBB2 (Young et al., 2008).
Thus, it seems plausible to hypothesize that the elevated ERBB2 expression or activity frequently observed during acquired resistance to endocrine therapy may result in heterodimerization with ERBB4, enabling ERBB4 to contribute to this acquired resistance (Ghayad et al., 2010). Indeed, as we have stated elsewhere, we predict that functional differences between ERBB4 homodimers (tumor suppressors) and heterodimers (oncoproteins) account for the disparities in ERBB4 function observed in breast cancers.
Functional differences among the ERBB4 isoforms may also account for the fact that ERBB4 appears to function as a tumor suppressor and as an oncoprotein, particularly since the JMb isoforms cannot be processed into the 4ICD intracellular fragment and thereby cannot facilitate the trafficking of signaling proteins to the nucleus and mitochondria. There is also evidence that the Cyt1 and Cyt2 isoforms play distinct roles in breast cancers (Fujiwara et al., 2014; Segers et al., 2020). This functional distinction is likely due to differences in the coupling of the carboxyl terminus of ERBB4 to signaling effectors (see Section II.H and Section II.I for further details). The functional differences between ERBB4 homodimers (tumor suppressors) and heterodimers (oncoproteins) may also account for this apparent difference in ERBB4 functions.
Mutations in ERBB4 are observed in 1%–4% of all breast cancers (Mishra et al., 2017). However, this particular report failed to identify the individual ERBB4 mutations. The ERBB4 E872K mutation was found in an unspecified breast cancer sample (Soung et al., 2006; Tvorogov et al., 2009). As will be discussed later, it has been reported that the E872K mutant exhibits a gain-of-function phenotype (Prickett et al., 2009). Based on the hypothesis that ERBB4 functions as both tumor suppressor gene and as an oncogene, we predict that ERBB4 loss-of-function mutants will be found in the context in which ERBB4 functions as a tumor suppressor protein and that ERBB4 gain-of-function mutants will be found in the context in which ERBB4 functions as an oncoprotein.
C. Melanoma
ERBB4 is mutated in 15 of 79 (19%) melanoma samples, accounting for 20 distinct mutations (L39F, Y111H, M313I, E317K, S341L, R393W, P409L, E452K, R491K, E542K, R544W, E563K, D609N, P700S, E836K, E872K, G936R, P1033S, R1174Q, and S1246N; Tables 1 and 2) (Prickett et al., 2009). The L39F, R393W, and E872K mutations have been described in other tumor types, suggesting that these mutations are functionally relevant (Soung et al., 2006; Prickett et al., 2009; Tvorogov et al., 2009; Chen et al., 2015). Moreover, seven ERBB4 missense mutants (E317K, E452K, E542K, R544W, E563K, E836K, and E872K) found in these samples exhibit increased ERBB4 tyrosine phosphorylation and ERBB4 tyrosine kinase activity. Moreover, these seven ERBB4 missense mutants induce foci (loss of contact inhibition) in a monolayer of NIH 3T3 fibroblasts. The pan-ErbB tyrosine kinase inhibitor N-{3-chloro-4-[(3-fluorobenzyl)oxy]phenyl}-6-[5-({[2-(methylsulfonyl)ethyl]amino}methyl)-2-furyl]-4-quinazolinamine (lapatinib) (Rusnak et al., 2001) inhibits the proliferation of melanoma cell lines that possess endogenous ERBB4 mutant alleles but not cells that possess wild-type ERBB4. The inhibition of proliferation by lapatinib is accompanied by reduced ERBB4 tyrosine phosphorylation but not reduced ERBB2 tyrosine phosphorylation, suggesting that reduced ERBB4 phosphorylation is the mechanism of this growth inhibition. Similarly, silencing endogenous ERBB4 in melanoma cells that possess an ERBB4 mutant results in growth arrest and reduced AKT phosphorylation. Together, these data have led to the conclusion that ERBB4 functions as an oncogene in melanoma (Kurppa and Elenius, 2009; Prickett et al., 2009; Easty et al., 2011; Kunz, 2013, 2014, 2015; Mishra et al., 2017) and that gain-of-function ERBB4 mutations are one of the multiple mechanisms by which melanoma cells activate the PI3K pathway (Kurppa and Elenius, 2009). Moreover, given that lapatinib is a much more potent inhibitor of ERBB2 tyrosine kinase activity than of ERBB4 tyrosine kinase activity (Rusnak et al., 2001; Qiu et al., 2008), lapatinib inhibition of proliferation by ERBB4-mutant melanoma cell lines suggests that ERBB4-ERBB2 heterodimerization and ERBB4 phosphorylation by ERBB2 is responsible for PI3K activation and deregulated proliferation in melanoma cells that harbor gain-of-function ERBB4 mutants.
On the surface, the conclusion that these gain-of-function ERBB4 melanoma mutants function as oncogenes contradicts evidence that the synthetic gain-of-function Q646C and I658E ERBB4 mutants function as tumor suppressor genes (Penington et al., 2002; Williams et al., 2003; Gallo et al., 2006, 2013; Pitfield et al., 2006; Vidal et al., 2007; Mill et al., 2011a). However, recall that we have postulated that ERBB4 heterodimers function as oncoproteins, whereas homodimers function as tumor suppressors. Thus, one possible explanation for the apparent difference between the gain-of-function ERBB4 melanoma mutants and the synthetic gain-of-function ERBB4 mutants is that Q646C is constitutively homodimerized and I658E is presumed to be constitutively homodimerized. In contrast, the gain-of-function ERBB4 melanoma mutants may cause increased heterodimerization with EGFR or ERBB2 in addition to or instead of increased ERBB4 homodimerization.
Another critique of these gain-of-function ERBB4 melanoma mutants is that they have not been useful in directing the use of lapatinib in a clinical trial for the treatment of advanced melanoma (Gonzalez-Cao et al., 2015; Mishra et al., 2017; Rudloff and Samuels, 2010). One potential explanation is that no group has replicated the finding that the E317K, E452K, E542K, R544W, E563K, E836K, and E872K ERBB4 mutants exhibit increased ligand-dependent or -independent phosphorylation nor the finding that these mutants cause loss of contact inhibition (focus formation) in a fibroblast cell line. Given that we postulate that gain-of-function ERBB4 melanoma mutants cause increased signaling by ERBB4 heterodimers but not by ERBB4 homodimers, differences in EGFR and ERBB2 expression in model systems could account for the apparent failure to replicate initial findings.
Another complication is that subsequent studies of melanoma genomes have either failed to detect the presumed seven gain-of-function ERBB4 alleles or have detected ERBB4 mutations at a greatly reduced frequency in melanoma (Manca et al., 2013; Zhou et al., 2013; Segers et al., 2020). On the other hand, after in silico filtering for functional relevance, ERBB4 is mutated in 12 of 100 (12%) melanoma samples, and 18 missense mutant alleles (16 distinct alleles) are found in these samples. Functional analyses of these mutants have not been performed. Only 2 of these 16 alleles correspond to the gain-of-function ERBB4 melanoma mutants described in the prior analysis of melanoma samples (E452K and E872K) (Prickett et al., 2009; de Unamuno Bustos et al., 2017). Similarly, these mutations are distributed across the entire ERBB4 coding sequence, and this study failed to reveal the profound ERBB4 mutational hot spots characteristic of many gain-of-function mutant alleles in oncogenes, including gain-of-function mutations in EGFR, BRAF, and the neuroblastoma RAS viral oncogene homolog (NRAS) gene. Thus, it is possible that at least some of these 16 ERBB4 mutations cause a loss of function in the context of a homodimeric ERBB4 tumor suppressor (de Unamuno Bustos et al., 2017). Indeed, there have been calls for additional studies into the mechanisms by which ERBB4 melanoma mutants drive malignancies, in apparent recognition of the possibility that some ERBB4 melanoma mutants drive malignancies by disrupting the activity of homodimeric ERBB4 tumor suppressors (Rudloff and Samuels, 2010; Mishra et al., 2017).
VI. Implications for Staging and Treating Human Tumors
A. Determination of whether ERBB4 Functions as a Tumor Suppressor or as an Oncoprotein
Our hypothesis that ERBB4 homodimers function as tumor suppressors, whereas ERBB4 heterodimers possess oncogenic activity, has profound implications the potential utility of ERBB4 as a biomarker and therapeutic target (Fig. 7). The most profound implication is the obvious need to assess the contribution of ERBB4 to the malignant phenotype for each tumor type of interest. Multiple approaches will need to be deployed.
A proposed role for ERBB4 in human malignancies.
Relative to normal samples, ERBB4 overexpression or elevated ERBB4 copy number in tumor samples will be a preliminary indication that ERBB4 functions as an oncogene in those samples. Because ERBB4 heterodimers are oncogenic, ERBB4 may also function as an oncogene in some EGFR-dependent or ERBB2-dependent tumors. These hypotheses will need to be confirmed through overexpression studies in normal tissues to confirm that ERBB4 is sufficient for malignant growth transformation. The hypothesis will also need to be confirmed through knockdown/silencing studies in tumor cells to confirm that ERBB4 is necessary for malignant phenotypes.
Relative to normal samples, ERBB4 underexpression or loss of ERBB4 copy number in tumor samples will be a preliminary indication that ERBB4 functions as a tumor suppressor gene in those samples. Such a hypothesis will need to be confirmed through knockdown/silencing studies in normal cells to demonstrate that loss of ERBB4 expression is sufficient for malignant growth transformation. The hypothesis will also need to be confirmed through overexpression studies in tumor cells to demonstrate that loss of ERBB4 expression is necessary for malignant phenotypes.
B. Identification of ERBB4 Mutants that Serve as Biomarkers through Their Function as Bona Fide Tumor Drivers
As noted in Table 1 and elsewhere, ERBB4 polymorphisms (most notably, missense mutations) are found in a wide variety of human malignancies. Strategies for ascertaining the functional significance of particular ERBB4 missense mutations in a specific type of tumor must account for the possibility that ERBB4 functions either as an oncogene or as a tumor suppressor gene in that particular tumor type. Thus, the general question of whether ERBB4 functions as a tumor suppressor gene or as an oncogene in a specific type of tumor must precede the determination of whether particular ERBB4 missense mutations function as bona fide tumor drivers.
In tumors in which ERBB4 functions as an oncogene, ERBB4 missense mutations that act as bona fide tumor drivers are anticipated to exhibit a gain-of-function phenotype. In other words, relative to wild-type ERBB4, the ERBB4 missense mutants will encode proteins that exhibit elevated tyrosine phosphorylation and coupling to downstream signaling effectors. To demonstrate that these mutants function as bona fide tumor drivers, the expression of the ERBB4 missense mutants in normal cells must induce malignant phenotypes to a greater extent than does wild-type ERBB4. Furthermore, knockdown/silencing of endogenous ERBB4 mutants in tumor cells must inhibit malignant phenotypes, and these malignant phenotypes must not be rescued through reintroduction of wild-type ERBB4 at endogenous levels of expression.
In tumors in which ERBB4 functions as a tumor suppressor, ERBB4 missense mutations that act as bona fide tumor drivers are anticipated to exhibit a loss-of-function phenotype. In other words, relative to wild-type ERBB4, the ERBB4 missense mutants will encode proteins that exhibit decreased tyrosine phosphorylation and coupling to downstream signaling effectors. To demonstrate that these mutants function as bona fide tumor drivers, the ERBB4 missense mutations must disrupt the growth inhibitory activity of wild-type ERBB4. In particular, knockdown/silencing of endogenous ERBB4 mutants in tumor cells must inhibit malignant phenotypes, and these malignant phenotypes must not be rescued through reintroduction of wild-type ERBB4.
Note that we predict that ERBB4 gain-of-function tumor driver mutations will potentiate the phosphorylation and oncogenic signaling of ERBB4 heterodimers to a greater extent than they will potentiate the phosphorylation and tumor suppressor signaling of ERBB4 homodimers. Likewise, we predict that ERBB4 loss-of-function tumor driver mutations will disrupt the phosphorylation and tumor suppressor signaling by ERBB4 homodimers to a greater extent than they will disrupt oncogenic signaling by ERBB4 heterodimers. Therefore, significant forethought must go into the choice of model systems used to evaluate the activity of ERBB4 mutants, particularly with respect to the expression of other ERBB family receptors.
Likewise, the characterization of ERBB4 mutants must avoid using ERBB4 tyrosine phosphorylation as the sole indication of ERBB4 signaling activity. As noted elsewhere, the synthetic ERBB4 Q646C mutant exhibits ligand-independent homodimerization, tyrosine phosphorylation, and tumor suppressor activity. On the other hand, the synthetic ERBB4 H647C and A648C mutants exhibit ligand-independent homodimerization and tyrosine phosphorylation but do not possess tumor suppressor activity. It has been hypothesized that the H647C and A648C mutants do not homodimerize in the same conformation as the Q646C mutant, resulting in differences in the sites of ERBB4 tyrosine phosphorylation and differential coupling to effectors and growth inhibitory signaling (Penington et al., 2002; Williams et al., 2003; Pitfield et al., 2006).
C. Potential Therapeutic Approaches for Tumors that Harbor Tumor Driver Mutations in ERBB4
The use of ERBB4 tumor driver mutants as biomarkers for therapeutic intervention must reflect whether such mutants potentiate the oncogenic activity of ERBB4 or disrupt the tumor suppressor activity of ERBB4. This is because ERBB4 itself is not considered druggable in contexts in which it functions as a homodimeric tumor suppressor. In contrast, the wealth of United States Food and Drug Administration–approved agents that target EGFR and ERBB2 suggest that ERBB4 is quite druggable in contexts in which it functions as a heterodimeric oncoprotein. Some small-molecule inhibitors for other kinases nonetheless have reasonable potency against ERBB4 (Qiu et al., 2008; Ahammad et al., 2020). Moreover, classic medicinal chemistry approaches could be employed to improve the selectivity of these existing inhibitors for ERBB4.
On the other hand, here we propose that the observation that ERBB4 kinase activity is dispensable for the growth stimulatory activity of ERBB4 heterodimers (Pitfield et al., 2006; Mill et al., 2011a,b; Wilson et al., 2012b) is generally applicable to tumors in which ERBB4 heterodimers function as oncoproteins. If this holds true, then agents that disrupt EGFR or ERBB2 signaling possess more promise for the treatment of tumors that are dependent on ERBB4 heterodimers than small-molecule ERBB4 tyrosine kinase inhibitors. Indeed, inhibiting ERBB4 homodimeric signaling by small-molecule ERBB4 tyrosine kinase inhibitors is predicted to be deleterious.
ERBB4 mutants that function as bona fide tumor drivers by potentiating signaling by oncogenic ERBB4-EGFR or ERBB4-ERBB2 heterodimers may indicate sensitivity to inhibitors of effectors of these ERBB4 heterodimers. As discussed earlier, the PI3K/AKT pathway appears to be a frequent mediator of oncogenic ERBB4 signaling. Section II.D describes other ERBB4 effectors that may be appealing targets for therapy.
It is much less apparent how to treat tumors that harbor ERBB4 (loss-of-function) tumor driver mutations that disrupt the tumor suppressor activity of ERBB4 homodimers. Section II.H describes mechanisms of homotypic ERBB4 signaling through the 4ICD, including mechanisms that lead to apoptosis and cell cycle arrest. Therefore, in tumors in which 4ICD signaling is lost due to a loss-of-function tumor driver mutation in ERBB4, therapeutics that promote apoptosis or cell cycle arrest may be effective.
Acknowledgments
The authors thank Profs. David Stern and Mark Lemmon of the Yale Cancer Center for decades of insightful conversations regarding ERBB4 function.
Authorship Contributions
Wrote or contributed to the writing of the manuscript: Lucas, Dwivedi, Senfeld, Cullum, Mill, Piazza, Bryant, Cook, Miller, Lott, Kelley, Knerr, Markham, Kaufmann, Jacobi, Shen, Riese.
Footnotes
- Received May 14, 2021.
- Accepted August 15, 2021.
Research in the Riese laboratory has been funded by an Auburn University Research Initiative in Cancer (AURIC) fellowship to R.L.C., an AURIC grant to D.J.R., a US Department of Education GAANN Graduate Fellowship Program in Biological & Pharmaceutical Engineering Award P200A120244 to Auburn University, an Auburn University Presidential Graduate Research Fellowship to L.M.L., an Auburn University Intramural Grants Program award to D.J.R., and other support from the Auburn University Department of Drug Discovery and Development and the Auburn University Harrison School of Pharmacy.
D.J.R. is a consultant for Eli Lilly and Company, Bristol-Myers Squibb, and ImClone, LLC. R.L.C. is an employee of Abbott Laboratories. The funders had no role in preparation of the manuscript.
Abbreviations
- ADAM17
- TNF converting enzyme
- AKT
- protein kinase
- ALCL
- anaplastic large-cell lymphoma
- AREG
- amphiregulin
- ATM
- ataxia telangiectasia mutated
- BAK
- Bcl-2 homologous antagonist/killer
- BAPC
- anaphase-promoting complex
- BAX
- Bcl-2-associated X protein
- BCBL
- Casitas B-lineage lymphoma E3 ubiquitin ligase
- BH3
- Bcl2 homology 3 domain
- BRAF
- v-Raf murine sarcoma viral oncogene homolog
- BTC
- betacellulin
- CRC
- colorectal cancer
- CT
- carboxyl terminal
- Cyt1/Cyt2
- cytoplasmic domain isoforms of ERBB4 that arise through alternative splicing of the ERBB4 transcript
- ECD
- extracellular subdomain of ERBB4
- EGF
- epidermal growth factor
- EGFR
- ERBB1
- ER
- estrogen receptor
- G2/M
- transition between the G2 and M phases of the cell cycle
- GRB
- growth factor receptor-bound
- HBEGF
- heparin-binding EGF-like growth factor
- HCC
- hepatocellular carcinoma
- HPV
- human papillomavirus
- HNSCC
- head and neck squamous cell carcinoma
- 4ICD
- soluble form of the ERBB4 cytoplasmic (intracellular) domain
- ITCH
- itchy E3 ubiquitin protein ligase
- JMa/JMb
- extracellular juxtamembrane isoforms of ERBB4 that arise through alternative splicing of the ERBB4 transcript
- KRAS
- Kirsten RAS
- MDM2
- mouse double minute 2 homolog
- miR
- microRNA
- MMTV
- mouse mammary tumor virus promoter
- NRG
- neuregulin
- OMM
- mitochondrial outer membrane
- p85
- regulatory subunit of phosphatidylinositol 3-kinase
- PAY
- proline-proline-alanine-tyrosine
- PDAC
- pancreatic ductal adenocarcinoma
- PDZ
- structural motif first described in postsynaptic density protein 95 (PSD-95), Dlg1, and Zo-1 and involved in interactions of membrane-bound receptors with cytoskeletal proteins
- PI3K
- phosphatidylinositol 3-kinase
- PKB
- protein kinase BP
- PR
- progesterone receptor
- RAS
- rat sarcoma virus
- RIP
- regulated intramembrane proteolysis
- SRC
- Rous sarcoma virus protein tyrosine kinase
- SKCM
- skin cutaneous melanoma
- STAD
- stomach adenocarcinoma
- STAT
- signal transducer and activator of transcription
- TCGA
- The Cancer Genome Atlas
- TGFA
- transforming growth factor α
- TNBC
- triple-negative breast cancer
- TNF
- tumor necrosis factor
- WWOX
- tumor suppressor oxidoreductase
- YAP1
- Yes-associated protein 1
- Copyright © 2022 by The Author(s)
This is an open access article distributed under the CC BY-NC Attribution 4.0 International license.
References
- ↵
- ↵
- ↵
- ↵
- ↵
- ↵
- ↵
- ↵
- ↵
- ↵
- ↵
- ↵
- ↵
- ↵
- ↵
- ↵
- ↵
- ↵
- ↵
- ↵
- ↵
- ↵
- ↵
- ↵
- ↵
- ↵
- ↵
- ↵
- ↵
- ↵
- ↵
- ↵
- ↵
- ↵
- ↵
- ↵
- ↵
- ↵
- ↵
- ↵
- ↵
- ↵
- ↵
- ↵
- ↵
- ↵
- ↵
- ↵
- ↵
- ↵
- ↵
- ↵
- ↵
- ↵
- ↵
- ↵
- ↵
- ↵
- ↵
- ↵
- ↵
- ↵
- ↵
- ↵
- ↵
- ↵
- ↵
- ↵
- ↵
- ↵
- ↵
- ↵
- ↵
- ↵
- ↵
- ↵
- ↵
- ↵
- ↵
- ↵
- ↵
- ↵
- ↵
- ↵
- ↵
- ↵
- ↵
- ↵
- ↵
- ↵
- ↵
- ↵
- ↵
- ↵
- ↵
- ↵
- ↵
- ↵
- ↵
- ↵
- ↵
- ↵
- ↵
- ↵
- ↵
- ↵
- ↵
- ↵
- ↵
- ↵
- ↵
- ↵
- ↵
- ↵
- ↵
- ↵
- ↵
- ↵
- ↵
- ↵
- ↵
- ↵
- ↵
- ↵
- ↵
- ↵
- ↵
- ↵
- ↵
- ↵
- ↵
- ↵
- ↵
- ↵
- ↵
- ↵
- ↵
- ↵
- ↵
- ↵
- ↵
- ↵
- ↵
- ↵
- ↵
- ↵
- ↵
- ↵
- ↵
- ↵
- ↵
- ↵
- ↵
- ↵
- ↵
- ↵
- ↵
- ↵
- ↵
- ↵
- ↵
- ↵
- ↵
- ↵
- ↵
- ↵
- ↵
- ↵
- ↵
- ↵
- ↵
- ↵
- ↵
- ↵
- ↵
- ↵
- ↵
- ↵
- ↵
- ↵
- ↵
- ↵
- ↵
- ↵
- ↵
- ↵
- ↵
- ↵
- ↵
- ↵
- ↵
- ↵
- ↵
- ↵
- ↵
- ↵
- ↵
- ↵
- ↵
- ↵
- ↵
- ↵
- ↵
- ↵
- ↵
- ↵
- ↵
- ↵
- ↵
- ↵
- ↵
- ↵
- ↵
- ↵
- ↵
- ↵
- ↵
- ↵
- ↵
- ↵
- ↵
- ↵
- ↵
- ↵
- ↵
- ↵
- ↵
- ↵
- ↵
- ↵
- ↵
- ↵
- ↵
- ↵
- ↵
- ↵
- ↵
- ↵
- ↵
- ↵
- ↵
- ↵
- ↵
- ↵
- ↵
- ↵
- ↵
- ↵
- ↵
- ↵
- ↵
- ↵
- ↵
- ↵
- ↵
- ↵
- ↵
- ↵
- ↵
- ↵
- ↵
- ↵
- ↵
- ↵
- ↵
- ↵
- ↵
- ↵
- ↵
- ↵
- ↵
- ↵
- ↵
- ↵
- ↵
- ↵
- ↵
- ↵
- ↵
- ↵
- ↵
- ↵
- ↵
- ↵
- ↵
- ↵
- ↵
- ↵
- ↵
- ↵
- ↵
- ↵
- ↵
- ↵
- ↵
- ↵
- ↵
- ↵
- ↵
- ↵
- ↵
- ↵
- ↵
- ↵
- ↵
- ↵
- ↵
- ↵
- ↵
- ↵
- ↵
- ↵
- ↵
- ↵
- ↵
- ↵
- ↵
- ↵
- ↵
- ↵
- ↵
- ↵
- ↵
- ↵
- ↵
- ↵
- ↵
- ↵
- ↵
- ↵
- ↵
- ↵
- ↵
- ↵
- ↵
- ↵
- ↵
- ↵
- ↵
- ↵
- ↵
- ↵
- ↵
- ↵
- ↵
- ↵
- ↵
- ↵
- ↵
- ↵
- ↵
- ↵
- ↵
- ↵
- ↵