Abstract
Background:
Currently, sunitinib represents one of the therapeutic strongholds for renal cell carcinoma, but the criteria for treatment selection are lacking. We assessed the role of vascular endothelial growth factor (VEGF) and VEGF receptor (VEGFR) polymorphisms in the prediction of the clinical outcome in metastatic renal cell carcinoma (mRCC) patients.
Methods:
A total of 84 tumour samples from mRCC patients receiving first-line sunitinib were tested for VEGF and VEGFR single-nucleotide polymorphisms (SNPs). The SNP results were correlated with progression-free survival (PFS) and overall survival (OS).
Results:
Median PFS was 8.22 months, although whereas median OS was 32.13 months. The VEGF A rs833061 resulted significant in PFS (17 vs 4 months; P<0.0001) and OS (38 vs 10 months; P<0.0001). The VEGF A rs699947 was significant for PFS (18 vs 4 months; P=0.0001) and OS (37 vs 16 months; P<0.0001). The VEGF A rs2010963 was significant in PFS (18 vs 8 vs 2 months; P=0.0001) and OS (31 vs 36 vs 9 months; P=0.0045). The VEGR3 rs6877011 was significant in PFS (12 vs 4 months; P=0.0075) and OS (36 vs 17 months; P=0.0001). At multivariate analysis, rs833061, rs2010963 and rs68877011 were significant in PFS, and rs833061 and rs68877011 were independent factors in OS.
Conclusions:
In our analysis, patients with TT polymorphism of rs833061, CC polymorphism of rs699947, CC polymorphism of rs2010963 and CG polymorphism of rs6877011 seem to have a worse PFS and OS when receiving first-line sunitinib.
Similar content being viewed by others
Main
The metastatic renal cell carcinoma (mRCC) therapy scenario has radically changed in recent years, and currently the therapeutic strongholds are mostly represented by tyrosine-kinase inhibitors (TKIs) directed against the vascular endothelial growth factor (VEGF) signalling pathway. One of these new molecules, approved for first-line mRCC treatment, is Sunitinib (Motzer et al, 2000; Lam et al, 2005).
Nevertheless, in spite of interesting activity profile, a large proportion of patients, ranging from 60 to 70%, are still refractory to sunitinib and, therefore, they are exposed to potentially relevant toxicities without any clinical benefit (Motzer et al, 2007, 2009).
Hypoxia and compensatory hyperactivation of angiogenesis are particularly important in RCC, given the highly vascularised nature of kidney tumours and the specific association of mutation in VHL, a critical regulator of the hypoxic response, with the onset of RCC (Pantuck et al, 2003).
The VEGF family members are secreted, dimeric glycoproteins of ∼40 kDa, consisting of five members, VEGF A, B, C, D and placental growth factor (PLGF), and binding to specific receptors (Valtola et al, 1999; Jia et al, 2004; Rini et al, 2008; Stuttfeld and Ballmer-Hofer, 2009; Zhang et al, 2009).
The VEGF gene is quite complex, with several alternatively spliced isoforms, and the regulation of expression could differ between normal and tumour tissue. Interestingly, as all identified polymorphisms in VEGF are not in the coding region, alternative mechanisms for their role in gene expression have been proposed. In fact, although many transcription factors bind to the promoter regions of VEGF (Pages and Puyssegur, 2005), none occur at the common polymorphic sites associated with VEGF expression. Nevertheless, single-nucleotide polymorphisms (SNPs) have been reported to cause changes in VEGF expression levels (Pander et al, 2007).
The SNPs in the VEGF and VEGF receptor (VEGFR) genes have been also correlated with tumour neoangiogenesis through different biological mechanisms.
Numerous SNPs in the promoter, 5′-, and 3′-untranslated regions (UTRs) are present in VEGF family genes. The 5′- and 3′-UTR contains key regulatory elements that are sensitive to hypoxia (Minchenko et al, 1994), and contribute to high variability in VEGF production among tissues (Vaziri et al, 2010). For example, 634 G>C SNP in the 5′-UTR of VEGF affects the protein translation efficiency (Schultz et al, 1999), and 936 C>T SNP in the 3′-UTR influences the circulating plasma concentrations (Watson et al, 2000) and tumour tissue expression of VEGF (Renner et al, 2000). However, it is likely that only a small number of these polymorphisms and haplotypes (linearly linked SNPs) actually have a functional effect on VEGF translation, whereas others act as proxies (Koukourakis et al, 2004).
Although a growing body of evidence suggested a possible correlation between an altered expression of the angiogenetic pathway and global outcome in colorectal, breast and ovarian patients treated with antiangiogenetic therapy (Schneider et al, 2008; Schultheis et al, 2008; Hansen et al, 2010; Steffensen et al, 2010; Hansen et al, 2011), data in mRCC are lacking. In a study performed on blood samples and tumour tissue specimens, Kim et al (2012) showed a statistically significant difference in patients with SNP −634 for sunitinib-related hypertension. Another study published by Garcia-Donas et al (2011) correlated SNPs with response and toxicities in mRCC patients treated with sunitinib. The authors showed that polymorphisms in VEGFR3 and CYP3A5*1 might be able to define a subset of patients with decreased sunitinib response and tolerability.
Based on these premises we evaluated the potential role of VEGF and VEGFR polymorphisms to define specific patients subgroups more likely to benefit from sunitinib therapy in terms of progression-free survival (PFS) and overall survival (OS).
Patients and methods
Patient selection
A total of 84 patients receiving first-line sunitinib treatment for histologically proven advanced renal cell carcinoma were eligible.
Follow-up consisted of physical examination, a complete blood count, chest radiography and US of the abdomen or CT/MRI scanning as clinically indicated.
The VEGF and VEGFR genotyping was performed on formalin-fixed, paraffin-embedded tissue block (∼30 mg) of renal cell carcinoma samples in nephrectomy or core biopsies, taken from the neoplasm periphery.
Paraffin wax was removed with xylene and the samples were washed twice with 100% ethanol. DNA was isolated from the deparaffinised tissue using the RecoverAll Total Nucleic Acid Isolation Kit for FFPE Tissues (Applied Biosystems, Foster City, CA, USA) according to the manufacturer’s instructions. DNA from each sample was then eluted in 120 μl of eluting solution.
The SNPs within each gene were selected using the Pupasuite software (http://pupasuite.bioinfo.cipf.es/index.jsf – version 2.0.0, bioinfo 2008), the CIPF SNP database (dbSNP) generated by the National Centre for Biotechnology Information (http://www.ncbi.nlm.nih.gov/SNP) and by review of the medical literature, using the following criteria:
-
1)
The polymorphism had some degree of likelihood to alter the structure or the expression of the gene in a biologically relevant manner (i.e., affecting ESE sequences, 3′-UTR or promoter region);
-
2)
The minor allele frequency was above 10% (with the only exception of rs2305948, rs6877011 and rs307822); and
-
3)
The genetic polymorphism was established and well documented.
Further considerations drove the selection of SNPs for our study. A correlation between the presence of a specific allele on a polymorphic site and the expression of the respective protein has been previously documented for VEGF (Formento et al, 2009; Chen et al, 2011). The SNPs in regulatory sequences, such as introns and 5′- and 3′-UTRs, have been shown to affect mRNA stability, processing efficiency, isoform expression and localisation. Moreover, regulatory motif sequences within the 3′ UTR of mRNAs have been shown to affect the stability of the messenger and/or its translational efficiency. Thus, it can be argued that SNPs in these sequences may influence VEGF and VEGFR gene expression. Also on these bases, we selected the SNPs known to affect VEGF and VEGFR expression and those located in regulatory sequences, for which a putative role in protein regulation can be assumed.
Globally, we assumed that selected SNPs had an impact on protein expression and therefore on biological function.
The selected SNPs were as follows: six polymorphisms in the VEGFA gene (rs10434, G>A; rs2010963, G>C; rs25648, C>T; rs3025039, C>T; rs699947, A>C; rs833061, C>T), two in VEGFC (rs4604006, T>C; rs7664413, C>T), two in VEGFR1 (FLT1) (rs664393, G>A; rs7993418, A>G), four in VEGFR2 (KDR) (rs1870377, A>T; rs2071559, A>G; rs2305948, G>A; rs7667298, A>G) and three in VEGFR3 (FLT4) (rs307805, A>G; rs6877011, C>G; rs307822, G>A). Chromosomal locations, positions and biological effects of investigated VEGF and VEGFR SNPs have been summarised in Table 1.
The SNP genotyping was performed by TaqMan technology using SNP genotyping products (Applied Biosystems). The PCR was performed and genotypes were analysed on the 7300 Real-Time PCR System (Applied Biosystems) using an ABI Prism 7300 Sequence Detection System software (version 1.3.1, Applied Biosystems). Each reaction contained 0.2 μl of total genomic DNA. Laboratory personnel blinded to patient status performed genotyping, and a random 10% of the samples were repeated to validate genotyping procedures.
All SNPs genotyped had to present an overall call rate of ≥90% to be included in our analysis; all samples resulted significant during the analysis and did not need test repetition.
Statistical analysis
Statistical analysis was performed with the MedCalc software version 10.4.8 (Mariakerke, Belgium) for Windows.
The association between categorical variables was estimated by the χ2 test.
Hazard ratios (HRs) for median PFS and OS between groups were estimated from Cox Regression models. The multivariate analysis also included adjustments for other variables such as age (≥65 vs <65 years), sex, performance status (Eastern Cooperative Oncology Group performance score, 0–1 vs 2), haemoglobin at the time of treatment start (less than lower limit of normal vs normal), lactate dehydrogenase (>1.5 × the upper limit of normal vs normal), corrected calcium (>10 vs <10 mg dl−1) and nephrectomy (yes vs not).
All polymorphisms were examined for deviation from Hardy–Weinberg equilibrium using the Powermarker v. 3.25 package (http://statgen.ncsu.edu/powermarker).
Linkage disequilibrium (LD) analysis was also performed using the Powermarker v. 3.25 package (www.statgen.ncsu.edu/powermarker). The LD was estimated using r2, with r2=1 indicating complete LD and r2=0 indicating absent LD.
Results
Hardy–Weinberg equilibrium and LD
The frequencies of the tested genotypes resulted comparable to those reported in Caucasians, with no significant deviation from the Hardy–Weinberg equilibrium.
Linkage disequilibrium was observed for the tumour genotypes rs833061, rs699947 and rs2010963 of VEGF A (P>0.0001), correlated with either PFS or OS. No LD was observed for rs6877011 of VEGFR III.
Patient characteristics
For our analysis, 84 patients with histologically proven mRCC receiving first-line sunitinib were available: 65 males and 19 females with median age at diagnosis of 64 years (range 47–85) (Table 2).
In all, 73 patients underwent renal surgery, and for 11 patients, only core biopsies were available. Also, 29 patients were metastatic at diagnosis, and 21 of these patients were resectable. Of the patients, 77 had a clear cell renal cell carcinoma histology, and 7 had other types (2 sarcomatoid and 5 undefined). All patients received sunitinib as first-line treatment with standard schedule (4 weeks on/2 weeks off), and dose reduction was applied in patients with grade 3 and 4 toxicities, as clinically indicated.
No statistically significant differences were found according to genotype for major patients characteristics (PS, tumour burden, and so on).
In the general population, median PFS was 8.22 months, whereas median OS was 32.13 months.
Genotype analysis
All SNPs genotyped presented an overall call rate of ≥90%.
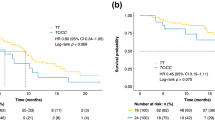
A total of 60 patients (71%) were found with a CC or CT genotype of rs833061, whereas 24 patients (29%) had TT genotype. Median PFS was improved for patients showing the CC/CT genotype (17 vs 4 months; P<0.0001; Figure 1), as also was median OS (38 vs 10 months; P<0.0001; Figure 2).
In all, 8 patients (10%) had a CC genotype of rs2010963, 39 patients (46%) had a CG genotype and 37 patients (44%) had a GG genotype. Progression-free survival proved statistically significant different among these genotypes with 2 months for CC genotype, 8 months for CG and 18 months for GG (P=0.0001; Figure 3). Overall survival was significant (9 vs 36 vs 31; P=0.0045).
A total of 60 patients (71%) had an AA or AC genotype for rs699947 and 24 patients (29%) had a CC genotype. Median PFS was improved for patients showing the AA/AC genotype (18 vs 4 months; P=0.0001), as also was median OS (37 vs 16 months; P<0.0001).
An analysis of rs833061, rs2010963 and rs699947 of VEGF A identifies a population of patients who express these polymorphisms in accordance to LD.
In all, 71 patients (85%) expressed a CC genotype of rs6877011 and 13 patients (15%) had a CG genotype. No patients expressed the GG genotype. For patients showing these polymorphisms, PFS was of 12 months for CC genotype and 4 months for CG genotype (P=0.0075; Figure 4) and OS was of 36 months for CC and 17 months for CG (P=0.0001; Figure 5 and Table 3).
On multivariate analysis, rs833061 (HR=0.71), rs2010963 (HR=0.19) and rs68877011 (HR=0.35) were significant in PFS. rs833061 (HR=0.69) and rs68877011 (HR=0.39) were also independent factors in OS.
Patients expressing all the favourable polymorphisms of rs833061, rs2010963, rs699947 and rs68877011 seem to have better overall response rate compared with those with unfavourable ones (56 vs 12%).
Discussion
Targeting the angiogenetic pathway resulted in a complete revolution in the treatment and prognosis of mRCC. However, in mRCC patients treated with anti-VEGF TKIs, PFS and OS may widely vary from patient to patient, ranging from few weeks to years, with no apparent explanation in most of the cases. These observations may not be easily connected to the previous known predictive and prognostic factors and risk categories (Pander et al, 2007).
A number of potential mechanisms of action involving both stromal and cancer cells have been hypothesised for sunitinib. Among these mechanisms, ‘vascular normalisation’ has the most robust clinical evidence (Jain, 2005). Deprimo et al. (2007) recently showed how TKIs targeting the RTKs (e.g., sunitinib) produce an increase in VEGF levels and a decrease in soluble VEGFR-2 (sVEGFR-2) and sVEGFR-3 in cytokine-refractory patients with mRCC. Interestingly, these changes in VEGF and sVEGFR were observed during treatment with sunitinib, and levels tended to return to near baseline after 2 weeks off treatment, indicating that these effects were dependent on drug exposure. Furthermore, significantly larger changes in VEGF, sVEGFR-2 and sVEGFR-3 levels were observed in patients exhibiting objective tumour response compared with those exhibiting stable disease or disease progression (P<0.05 for each analyte).
In our analysis, the CC+CT polymorphism of VEGF A rs833061 proved statistically significant in PFS (P<0.0001) and OS (P<0.0001) along with the AA+AC polymorphism of rs699947 (PFS, P=0.0001; OS, P<0.0001). The CC polymorphism of rs2010963 of VEGF A also showed significant correlation in PFS (P=0.0001) and OS (P=0.0045). rs833061 is located in the promoter region of the VEGF A gene on chromosome 6, similar to rs699947, whereas rs2010963 is located in the terminal 5′-UTR region of the VEGF A gene. We can hypothesise that different SNPs in different regions of the VEGF gene may influence circulating levels of VEGF and thus response to anti-VEGF therapies.
These findings and hypothesis could explain how a certain constitutive variation in VEGF and VEGFR levels could exert a significant difference in tumour outcome during antiangiogenetic treatment. Candidate gene studies exploring associations between VEGF polymorphisms and circulating VEGF levels have yielded controversial results. Eight studies have found significant associations with candidate polymorphisms (rs699947, rs1570360, rs833061, rs2010963, rs3025039, and 2549 18 bp I/D) in the promoter, 5′- and 3′-UTRs of the VEGF gene (Renner et al, 2000; Awata et al, 2002; Krippl et al, 2003; Ferrante et al, 2006; Zhai et al, 2007; Kamoun et al, 2008; Petrovic et al, 2008). However, several other studies did not identify any association with these and other VEGF SNPs. Using a hypothesis-free genome-wide approach, Debette et al (2011) found associations with 140 SNPs. Of these, 68 SNPs are located on chromosome 6, ∼150 kb downstream from the 3′ end of the VEGF gene, far from previously tested candidate SNPs. However, the real effect of SNPs in circulating or tumour tissue VEGF levels needs further studies in order to definitively associate a specific SNP to a specific effect on the corresponding growth factor or receptor.
In our population, we also found a statistical significance in PFS (P=0.0075) and OS (P=0.0001) in the CC polymorphism of rs6877011 of VEGFR 3.
Angiogenic sprouting involves specification of subpopulations of endothelial cells into tip cells that respond to VEGF guidance cues, and stalk cells that follow the tip cells and proliferate to form the vascular network (Gunningham et al, 2001). Recent evidence indicates that VEGF induces the membrane-bound Notch ligand delta-like 4 (Dll4) in the tip cells, which leads to the induction of the stalk cell phenotype in adjacent endothelial cells through activation of Notch-1 (Laakkonen et al, 2007).
In conclusion, our data show that by analysing polymorphisms of the VEGF and VEGFR genes, we could be able to select proper patients to be treated with sunitinib to improve treatment outcome. Further prospective studies are warranted to confirm our findings.
Change history
19 March 2013
This paper was modified 12 months after initial publication to switch to Creative Commons licence terms, as noted at publication
References
Awata T, Inoue K, Kurihara S, Ohkubo T, Watanabe M, Inukai K, Inoue I, Katayama S (2002) A common polymorphism in the 5′-untranslated region of the VEGF gene is associated with diabetic retinopathy in type 2 diabetes. Diabetes 51: 1635–1639
Chen MH, Tzeng CH, Chen PM, Lin JK, Lin TC, Chen WS, Jiang JK, Wang HS, Wang WS (2011) VEGF-406 T→ polymorphism and its association with VEGF expression and outcome to FOLFOX-4 treatment in patients with colorectal carcinoma. Pharmacogenomics J 11 (3): 227–236
Debette S, Visvikis-Siest S, Chen MH, Ndiaye NC, Song C, Destefano A, Safa R, Azimi Nezhad M, Sawyer D, Marteau JB, Xanthakis V, Siest G, Sullivan L, Pfister M, Smith H, Choi SH, Lamont J, Lind L, Yang Q, Fitzgerald P, Ingelsson E, Vasan RS, Seshadri S (2011) Identification of cis- and trans-acting genetic variants explaining up to half the variation in circulating vascular endothelial growth factor levels. Circ Res 109 (5): 554–563
Deprimo SE, Bello CL, Smeraglia J, Baum CM, Spinella D, Rini BI, Michaelson MD, Motzer RJ (2007) Circulating protein biomarkers of pharmacodynamic activity of sunitinib in patients with metastatic renal cell carcinoma: modulation of VEGF and VEGF-related proteins. J Transl Med 5: 32–43
Ferrante M, Pierik M, Henckaerts L, Joossens M, Claes K, Van Schuerbeek N, Vlietinck R, Rutgeerts P, Van Assche G, Vermeire S (2006) The role of vascular endothelial growth factor (VEGF) in inflammatory bowel disease. Inflamm Bowel Dis 12: 870–878
Formento JL, Etienne-Grimaldi MC, Francoual M, Pagès G, Onesto C, Formento P, Chamorey E, Dassonville O, Poissonnet G, Milano G (2009) Influence of the VEGF-A 936C>T germinal polymorphism on tumoral VEGF expression in head and neck cancer. Pharmacogenomics 10: 1277–1283
Garcia-Donas J, Esteban E, Leandro-García LJ, Castellano DE, del Alba AG, Climent MA, Arranz JA, Gallardo E, Puente J, Bellmunt J, Mellado B, Martínez E, Moreno F, Font A, Robledo M, Rodríguez-Antona C (2011) Single nucleotide polymorphism associations with response and toxic effects in patients with advanced renal-cell carcinoma treated with first-line sunitinib: a multicentre, observational, prospective study. Lancet Oncol 12 (12): 1143–1150
Gunningham SP, Currie MJ, Han C, Turner K, Scott PA, Robinson BA, Harris AL, Fox SB (2001) Vascular endothelial growth factor-B and vascular endothelial growth factor-C expression in renal cell carcinomas: regulation by the von Hippel-Lindau gene and hypoxia. Cancer Res 61: 3206–3211
Hansen TF, Garm Spindler KL, Andersen RF, Lindebjerg J, Brandslund I, Jakobsen A (2011) The predictive value of genetic variations in the vascular endothelial growth factor gene A in metastatic colorectal cancer. Pharmacogenomics J 11: 53–60
Hansen TF, Sørensen FB, Spindler KL, Olsen DA, Andersen RF, Lindebjerg J, Brandslund I, Jakobsen A (2010) Microvessel density and the association with single nucleotide polymorphisms of the vascular endothelial growth factor receptor 2 in patients with colorectal cancer. Virchows Arch 456: 251–260
Jain RK (2005) Normalization of tumor vasculature: an emerging concept in antiangiogenic therapy. Science 307: 58–62
Jia H, Bagherzadeh A, Bicknell R, Duchen MR, Liu D, Zachary I (2004) Vascular endothelial growth factor (VEGF)-D and VEGF-A differentially regulate KDR-mediated signaling and biological function in vascular endothelial cells. J Biol Chem 279: 36148–36157
Kamoun M, Houman MH, Hamzaoui A, Hamzaoui K (2008) Vascular endothelial growth factor gene polymorphisms and serum levels in Behcet’s disease. Tissue Antigen 72: 581–587
Kim JJ, Vaziri SAJ, Rini BI, Elson P, Garcia JA, Wirka R, Dreicer R, Ganapathi MK, Ganapathi R (2012) Association of VEGF and VEGFR2 single nucleotide polymorphisms with hypertension and clinical outcome in metastatic clear cell renal cell carcinoma patients treated with sunitinib. Cancer 118 (7): 1946–1954
Koukourakis MI, Papazoglou D, Giatromanolaki A, Bougioukas G, Maltezos E, Sivridis E (2004) VEGF gene sequence variation defines VEGF gene expression status and angiogenic activity in non-small cell lung cancer. Lung Cancer 46: 293–298
Krippl P, Langsenlehner U, Renner W, Yazdani-Biuki B, Wolf G, Wascher TC, Paulweber B, Haas J, Sammonigg H (2003) A common 936 C/T gene polymorphism of vascular endothelial growth factor is associated with decreased breast cancer risk. Int J Cancer 106: 468–471
Laakkonen P, Waltari M, Holopainen T, Takahashi T, Pytowski B, Steiner P, Hicklin D, Persaud K, Tonra JR, Witte L, Alitalo K (2007) Vascular endothelial growth factor receptor 3 is involved in tumor angiogenesis and growth. Cancer Res 67: 593–599
Lam JS, Lepert JT, Belldegrun AS, Figlin RA (2005) Novel approaches in the therapy of metastatic renal cell carcinoma. World J Urol 23: 202–212
Minchenko A, Salceda S, Bauer T, Caro T (1994) Hypoxia regulatory elements of the human vascular endothelial growth factor gene. Cell Mol Biol Res 40: 35–39
Motzer RJ, Hutson TE, Tomczak P, Michaelson MD, Bukowski RM, Oudard S, Negrier S, Szczylik C, Pili R, Bjarnason GA, Garcia-del-Muro X, Sosman JA, Solska E, Wilding G, Thompson JA, Kim ST, Chen I, Huang X, Figlin RA (2009) Overall survival and updated results for sunitinib compared with interferon alfa in patients with metastatic renal cell carcinoma. J Clin Oncol 27: 3584–3590
Motzer RJ, Hutson TE, Tomczak P, Michaelson MD, Bukowski RM, Rixe O, Oudard S, Negrier S, Szczylik C, Kim ST, Chen I, Bycott PW, Baum CM, Figlin RA (2007) Sunitinib versus interferon alfa in metastatic renal-cell carcinoma. N Engl J Med 356: 115–124
Motzer RJ, Murphy BA, Bacik J, Schwartz LH, Nanus DM, Mariani T, Loehrer P, Wilding G, Fairclough DL, Cella D, Mazumdar M (2000) Phase III trial of interferon alfa-2a with or without 13-cis-retinoic acid for patients with advanced renal cell carcinoma. J Clin Oncol 18: 2972–2980
Pages G, Puyssegur J (2005) Transcriptional regulation of the vascular endothelial growth factor gene - a concert of activating factors. Cardiovasc Res 65: 564–573
Pander J, Gelderblom H, Guchelaar HJ (2007) Pharmacogenetics of EGFR and VEGF inhibition. Drug Discov Today 12: 1054–1060
Pantuck AJ, Zeng G, Belldegrun AS, Figlin RA (2003) Pathobiology, prognosis and targeted therapy for renal cell carcinoma: exploiting the hypoxia-induced pathway. Clin Cancer Res 9: 4641–4652
Petrovic MG, Korosec P, Kosnik M, Osredkar J, Hawlina M, Peterlin B, Petrovic D (2008) Local and genetic determinants of vascular endothelial growth factor expression in advanced proliferative diabetic retinopathy. Mol Vis 14: 1382–1387
Renner W, Kotschan S, Hoffmann C, Obermayer-Pietsch B, Pilger E (2000) A common 936 C/T mutation in the gene for vascular endothelial growth factor is associated with vascular endothelial growth factor plasma levels. J Vasc Res 37: 443–448
Rini BI, Michaelson MD, Rosenberg JE, Bukowski RM, Sosman JA, Stadler WM, Hutson TE, Margolin K, Harmon CS, DePrimo SE, Kim ST, Chen I, George DJ (2008) Antitumor activity and biomarker analysis of sunitinib in patients with bevacizumab refractory metastatic renal cell carcinoma. J Clin Oncol 26: 3743–3748
Schneider BP, Wang M, Radovich M, Sledge GW, Badve S, Thor A, Flockhart DA, Hancock B, Davidson N, Gralow J, Dickler M, Perez EA, Cobleigh M, Shenkier T, Edgerton S, Miller KD (2008) Association of vascular endothelial growth factor and vascular endothelial growth factor receptor-2 genetic polymorphisms with outcome in trial of paclitaxel compared with paclitaxel compared with paclitaxel plus bevacizumab in advanced breast cancer: ECOG 2100. J Clin Oncol 20: 4672–4678
Schultheis AM, Lurje G, Rhodes KE, Zhang W, Yang D, Garcia AA, Morgan R, Gandara D, Scudder S, Oza A, Hirte H, Fleming G, Roman L, Lenz HJ (2008) Polymorphisms and clinical outcome in recurrent ovarian cancer treated with cyclophosphamide and bevacizumab. Clin Cancer Res 14: 7554–7563
Schultz A, Lavie L, Hochberg I, Beyar R, Stone T, Skorecki K, Lavie P, Roguin A, Levy AP, Steffensen KD, Waldstrom M, Brandslund I, Jakobsen A (1999) Interindividual heterogeneity in the hypoxic regulation of VEGF: significance for the development of the coronary artery collateral circulation. Circulation 100: 547–552
Steffensen KD, Waldstrom M, Brandslund I, Jakobsen A (2010) The relationship of VEGF polymorphisms with serum VEGF levels and progression-free survival in patients with epithelial ovarian cancer. Gynecol Oncol 117: 109–116
Stuttfeld E, Ballmer-Hofer K (2009) Structure and function of VEGF receptors. IUBMB Life 61 (9): 915–922
Valtola R, Salven P, Heikkila P, Taipale J, Joensuu H, Rehn M, Pihlajaniemi T, Weich H, de Waal R, Alitalo K (1999) VEGFR-3 and its ligand VEGF-C are associated with angiogenesis in breast cancer. Am J Pathol 154: 1381–1390
Vaziri SAJ, Kim J, Ganapathi MK, Ganapathi R (2010) Vascular endothelial growth factor polymorphisms: role in response and toxicity of tyrosine kinase inhibitors. Curr Oncol Rep 12: 102–108
Watson CJ, Webb NJ, Bottomley MJ, Brenchley PE (2000) Identification of polymorphisms within the vascular endothelial growth factor (VEGF) gene: correlation with variation in VEGF protein production. Cytokine 12: 1232–1235
Zhai R, Gong MN, Zhou W, Thompson TB, Kraft P, Su L, Christiani DC (2007) Genotypes and haplotypes of the VEGF gene are associated with higher mortality and lower VEGF plasma levels in patients with ARDS. Thorax 62: 718–722
Zhang F, Tang Z, Hou X, Lennartsson J, Li Y, Koch AW, Scotney P, Lee C, Arjunan P, Dong L, Kumar A, Rissanen TT, Wang B, Nagai N, Fons P, Fariss R, Zhang Y, Wawrousek E, Tansey G, Raber J, Fong GH, Ding H, Greenberg DA, Becker KG, Herbert JM, Nash A, Yla-Herttuala S, Cao Y, Watts RJ, Li X (2009) VEGF-B is dispensable for blood vessel growth but critical for their survival, and VEGF-B targeting inhibits pathological angiogenesis. Proc Natl Acad Sci USA 106: 6152–6157
Author information
Authors and Affiliations
Corresponding author
Additional information
This work is published under the standard license to publish agreement. After 12 months the work will become freely available and the license terms will switch to a Creative Commons Attribution-NonCommercial-Share Alike 3.0 Unported License.
Rights and permissions
From twelve months after its original publication, this work is licensed under the Creative Commons Attribution-NonCommercial-Share Alike 3.0 Unported License. To view a copy of this license, visit http://creativecommons.org/licenses/by-nc-sa/3.0/
About this article
Cite this article
Scartozzi, M., Bianconi, M., Faloppi, L. et al. VEGF and VEGFR polymorphisms affect clinical outcome in advanced renal cell carcinoma patients receiving first-line sunitinib. Br J Cancer 108, 1126–1132 (2013). https://doi.org/10.1038/bjc.2012.501
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/bjc.2012.501
Keywords
This article is cited by
-
Genetic polymorphisms as predictive biomarker of survival in patients with gastrointestinal stromal tumors treated with sunitinib
The Pharmacogenomics Journal (2018)
-
Prognostic and Predictive Factors for Renal Cell Carcinoma
Targeted Oncology (2018)
-
An exploratory association of polymorphisms in angiogenesis-related genes with susceptibility, clinical response and toxicity in gastrointestinal stromal tumors receiving sunitinib after imatinib failure
Angiogenesis (2017)
-
Angiogenesis genotyping and clinical outcome during regorafenib treatment in metastatic colorectal cancer patients
Scientific Reports (2016)
-
IL8 polymorphisms and overall survival in pazopanib- or sunitinib-treated patients with renal cell carcinoma
British Journal of Cancer (2015)