Abstract
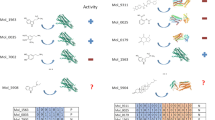
The clinical efficacy and safety of a drug is determined by its activity profile across many proteins in the proteome. However, designing drugs with a specific multi-target profile is both complex and difficult. Therefore methods to design drugs rationally a priori against profiles of several proteins would have immense value in drug discovery. Here we describe a new approach for the automated design of ligands against profiles of multiple drug targets. The method is demonstrated by the evolution of an approved acetylcholinesterase inhibitor drug into brain-penetrable ligands with either specific polypharmacology or exquisite selectivity profiles for G-protein-coupled receptors. Overall, 800 ligand–target predictions of prospectively designed ligands were tested experimentally, of which 75% were confirmed to be correct. We also demonstrate target engagement in vivo. The approach can be a useful source of drug leads when multi-target profiles are required to achieve either selectivity over other drug targets or a desired polypharmacology.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Hughes, J. D. et al. Physicochemical drug properties associated with in vivo toxicological outcomes. Bioorg. Med. Chem. Lett. 18, 4872–4875 (2008)
Roth, B. L. Drugs and valvular heart disease. N. Engl. J. Med. 356, 6–9 (2007)
Campillos, M., Kuhn, M., Gavin, A. C., Jensen, L. J. & Bork, P. Drug target identification using side-effect similarity. Science 321, 263–266 (2008)
Roth, B. L., Sheffler, D. J. & Kroeze, W. K. Magic shotguns versus magic bullets: selectively non-selective drugs for mood disorders and schizophrenia. Nature Rev. Drug Discov. 3, 353–359 (2004)
Knight, Z. A., Lin, H. & Shokat, K. M. Targeting the cancer kinome through polypharmacology. Nature Rev. Cancer 10, 130–137 (2010)
Brötz-Oesterhelt, H. & Brunner, N. A. How many modes of action should an antibiotic have? Curr. Opin. Pharmacol. 8, 564–573 (2008)
Hopkins, A. L. Network pharmacology: the next paradigm in drug discovery. Nature Chem. Biol. 4, 682–690 (2008)
Morphy, R. & Rankovic, Z. Designed multiple ligands. An emerging drug discovery paradigm. J. Med. Chem. 48, 6523–6543 (2005)
Paolini, G. V., Shapland, R. H. B., van Hoorn, W. P., Mason, J. S. & Hopkins, A. L. Global mapping of pharmacological space. Nature Biotechnol. 24, 805–815 (2006)
Keiser, M. J. et al. Relating protein pharmacology by ligand chemistry. Nature Biotechnol. 25, 197–206 (2007)
Keiser, M. J. et al. Predicting new molecular targets for known drugs. Nature 462, 175–181 (2009)
Vidal, D. & Mestres, J. In silico receptorome screening of antipsychotic drugs. Mol. Inf. 29, 543–551 (2010)
Lounkine, E. et al. Large-scale prediction and testing of drug activity on side-effect targets. Nature 486, 361–367 (2012)
Schneider, G. & So, S.-S. Adaptive Systems in Drug Design (Landes Biosciences, 2002)
Schneider, G. et al. Voyages to the (un)known: adaptive design of bioactive compounds. Trends Biotechnol. 27, 18–26 (2009)
Schneider, G., Lee, M. L., Stahl, M. & Schneider, P. De novo design of molecular architectures by evolutionary assembly of drug-derived building blocks. J. Comput. Aided Mol. Des. 14, 487–494 (2000)
Gillet, V. J., Willett, P., Fleming, P. J. & Green, D. V. Designing focused libraries using MoSELECT. J. Mol. Graph. Model. 20, 491–498 (2002)
Brown, N., McKay, B. & Gasteiger, J. The de novo design of median molecules within a property range of interest. J. Comput. Aided Mol. Des. 18, 761–771 (2004)
Nicolaou, C. A., Brown, N. & Pattichis, C. S. Molecular optimization using computational multi-objective methods. Curr. Opin. Drug Discov. Devel. 10, 316–324 (2007)
Liu, Q., Masek, B., Smith, K. & Smith, J. Tagged fragment method for evolutionary structure-based de novo lead generation and optimization. J. Med. Chem. 50, 5392–5402 (2007)
Dey, F. & Caflisch, A. Fragment-based de novo ligand design by multiobjective evolutionary optimization. J. Chem. Inf. Model. 48, 679–690 (2008)
Vinkers, H. M. et al. SYNOPSIS: SYNthesize and OPtimize system in silico. J. Med. Chem. 46, 2765–2773 (2003)
Heikkilä, T. et al. The first de novo designed inhibitors of Plasmodium falciparum dihydroorotate dehydrogenase. Bioorg. Med. Chem. Lett. 16, 88–92 (2006)
Roche, O. & Rodríguez Sarmiento, R. M. A new class of histamine H3 receptor antagonists derived from ligand based design. Bioorg. Med. Chem. Lett. 17, 3670–3675 (2007)
Alig, L. et al. Benzodioxoles: novel cannabinoid-1 receptor inverse agonists for the treatment of obesity. J. Med. Chem. 51, 2115–2127 (2008)
Schneider, G. et al. Reaction-driven de novo design, synthesis and testing of potential type II kinase inhibitors. Future Med. Chem. 3, 415–424 (2011)
Wermuth, C. G. Selective optimization of side activities: the SOSA approach. Drug Discov. Today 11, 160–164 (2006)
Gaulton, A. et al. ChEMBL: a large-scale bioactivity database for drug discovery. Nucleic Acids Res. 40, D1100–D1107 (2012)
Ribeiz, S. R. et al. Cholinesterase inhibitors as adjunctive therapy in patients with schizophrenia and schizoaffective disorder: a review and meta-analysis of the literature. CNS Drugs 24, 303–317 (2010)
Arnsten, A. F., Murphy, B. & Merchant, K. The selective dopamine D4 receptor antagonist, PNU-101387G, prevents stress-induced cognitive deficits in monkeys. Neuropsychopharmacology 23, 405–410 (2000)
Gillet, V., Johnson, A. P., Mata, P., Sike, S. & Williams, P. SPROUT: a program for structure generation. J. Comput. Aided Mol. Des. 7, 127–153 (1993)
Stahl, M. et al. A validation study on the practical use of automated de novo design. J. Comput. Aided Mol. Des. 16, 459–478 (2002)
Brown, N., McKay, B., Gilardoni, F. & Gasteiger, J. A graph-based genetic algorithm and its application to the multiobjective evolution of median molecules. J. Chem. Inf. Comput. Sci. 44, 1079–1087 (2004)
Nicolaou, C. A., Apostolakis, J. & Pattichis, C. S. De novo drug design using multiobjective evolutionary graphs. J. Chem. Inf. Model. 49, 295–307 (2009)
Stewart, K. D., Shiroda, M. & James, C. A. Drug Guru: a computer software program for drug design using medicinal chemistry rules. Bioorg. Med. Chem. 14, 7011–7022 (2006)
Kryssanov, V. V., Tamaki, H. & Kitamura, S. Understanding design fundamentals: how synthesis and analysis drive creativity, resulting in emergence. Artif. Intell. Eng. 15, 329–342 (2001)
Deb, K., Sundar, J., Udaya Bhaskara Rao, N. & Chaudhuri, S. Reference point based multi-objective optimization using evolutionary algorithms. Int. J. Comp. Intell. Res. 2, 273–286 (2006)
Lipinski, C. A., Lombardo, F., Dominy, B. W. & Feeney, P. J. Experimental and computational approaches to estimate solubility and permeability in drug discovery and development settings. Adv. Drug Deliv. Rev. 23, 3–25 (1997)
Ertl, P. & Schuffenhauer, A. Estimation of synthetic accessibility score of drug-like molecules based on molecular complexity and fragment contributions. J. Cheminform. 10, 8 (2009)
Fanelli, F. & De Benedetti, P. G. in Antitargets: Prediction and Prevention of Drug Side Effects (eds Vaz, R. J. & Klabunde, T. ) Ch. 8, 155–193 (Wiley-VCH, 2008)
Bemis, G. W. & Murcko, M. A. The properties of known drugs. 1. molecular frameworks. J. Med. Chem. 39, 2887–2893 (1996)
Ortega, R. et al. Synthesis, binding affinity and SAR of new benzolactam derivatives as dopamine D3 receptor ligands. Bioorg. Med. Chem. Lett. 19, 1773–1778 (2009)
Löber, S., Hübner, H., Tschammer, N. & Gmeiner, P. Recent advances in the search for D3 and D4 selective drugs: probes, models and candidates. Trends Pharmacol. Sci. 32, 148–157 (2011)
Martin, R. E., Green, L. G., Guba, W., Kratochwil, N. & Christ, A. Discovery of the first nonpeptidic, small-molecule, highly selective somatostatin receptor subtype 5 antagonists: a chemogenomics approach. J. Med. Chem. 50, 6291–6294 (2007)
Bender, A. et al. Chemogenomic data analysis: prediction of small-molecule targets and the advent of biological fingerprint. Comb. Chem. High Throughput Screen. 10, 719–731 (2007)
Rogers, D., Brown, R. D. & Hahn, M. Using extended-connectivity fingerprints with Laplacian-modified Bayesian analysis in high-throughput screening follow-up. J. Biomol. Screen. 10, 682–686 (2005)
Huang, X. P., Mangano, T., Hufeisen, S., Setola, V. & Roth, B. L. Identification of human Ether-à-go-go related gene modulators by three screening platforms in an academic drug-discovery setting. Assay Drug Dev. Technol. 8, 727–742 (2010)
Ruda, G. F. et al. Aryl phosphoramidates of 5-phosho erythronohydroxamic acid, a new class of potent trypanocidal agents. J. Med. Chem. 53, 6071–6078 (2010)
Pogorelov, V. M., Rodriguiz, R. M., Insco, M. L., Caron, M. G. & Wetsel, W. C. Novelty seeking and stereotypic activation of behavior in mice with disruption of the Dat1 gene. Neuropsychopharmacology. 30, 1818–1831 (2005)
Porton, B. et al. Mice lacking synapsin III show abnormalities in explicit memory and conditioned fear. Genes Brain Behav. 9, 257–268 (2010)
Glick, M., Jenkins, J. L., Nettles, J. H., Hitchings, H. & Davies, J. W. Enrichment of high-throughput screening data with increasing levels of noise using support vector machines, recursive partitioning, and laplacian-modified naive bayesian classifiers. J. Chem. Inf. Model. 46, 193–200 (2006)
Rogers, D. & Hahn, M. Extended-connectivity fingerprints. J. Chem. Inf. Model. 50, 742–754 (2010)
Truchon, J. F. & Bayly, C. I. Evaluating virtual screening methods: good and bad metrics for the “early recognition” problem. J. Chem. Inf. Model. 47, 488–508 (2007)
Zhao, W., Hevener, K. E., White, S. W., Lee, R. E. & Boyett, J. M. A statistical framework to evaluate virtual screening. BMC Bioinformatics 10, 225 (2009)
Cannon, E. O., Nigsch, F. & Mitchell, J. B. A novel hybrid ultrafast shape descriptor method for use in virtual screening. Chem. Cent. J. 2, 3 (2008)
Corne, D. W. & Knowles, J. D. in Proc. 9th Annual Conf. Genetic Evolutionary Computation 773–780 (ACM, 2007)
Obrezanova, O., Csanyi, G., Gola, J. M. & Segall, M. D. Gaussian processes: a method for automatic QSAR modelling of ADME properties. J. Chem. Inf. Model. 47, 1847–1857 (2007)
Obrezanova, O., Gola, J. M. R., Champness, E. J. & Segall, M. D. Automatic QSAR modeling of ADME properties: blood-brain barrier penetration and aqueous solubility. J. Comput. Aided Mol. Des. 22, 431–440 (2008)
Acknowledgements
This work is supported by SULSA (HR07019), the BBSRC Doctoral Training Programme, the BBSRC Pathfinder (BB/FOF/PF/15/09) and the BBSRC Follow On Fund schemes (BB/J010510/1) (A.L.H.), the University of Dundee’s Pump Priming Fund for Translational Medical Research (I.H.G. and A.L.H.) and by grants from the National Institutes of Health (NIH) supporting drug discovery receptor pharmacology (B.L.R.) and the NIH grant MH082441 (W.C.W.). The chemical synthesis and informatics benefits from the infrastructure investments from the Wellcome Trust Strategic Award (WT 083481). We thank J. Overington for StARlite and ChEMBL. We wish to thank D. Murugesan for compound purification and C. Means and T. Rhodes for helping with the open field, hole-board and zero-maze tests. We also wish to thank C. Elms and J. Zhou for their support in the husbandry and generation of the mice used for behavioural testing. We also wish to thank F. Y. Li for customizing the software configuration for the hole-board tests. Some of the equipment used in the behavioural testing was purchased with a grant from the North Carolina Biotechnology Center. B.L.R. also received support from the Michael Hooker Chair of Pharmacology.
Author information
Authors and Affiliations
Contributions
A.L.H. devised the method, developed the algorithm and designed the study. J.B. coded the algorithm and undertook the calculations. G.R.B. developed the databases. A.L.H. and J.B. with I.H.G., G.F.R. and K.A. selected the compounds for synthesis. I.H.G., G.F.R. and K.A. designed the synthetic routes and G.F.R. and K.A. undertook the chemical synthesis. L.A.W. purified and analysed several of the compounds. B.L.R. and V.S. designed the empirical tests for the synthesized compound predictions, analysed and interpreted the results and performed the experiments. X.-P.H. performed the 5-HT2B functional assays and the hERG assays. M.F.S. conducted the dopamine D2 and D4 functional assays. K.D.R. designed the drug metabolism and pharmacokinetics studies and analysed the results. S.N., L.S. and F.R.C.S. carried out the DMPK experiments. For the behavioural experiments, D.B.C. created the mice in which the Pcsk7 gene was disrupted. A.P. and N.G.S. verified the Pcsk7 deletion in many tissues including brain, and then backcrossed the mice onto a C57BL/6 background. W.C.W. designed the studies; R.M.R. and A.I.S. conducted the experiments and analysed the results; W.C.W., R.M.R., A.I.S., D.B.C., A.P. and N.G.S. interpreted the findings; A.L.H. and B.L.R. wrote the manuscript; I.H.G. wrote the synthetic methods with help from G.F.R., K.A. and L.A.W.; W.C.W. and R.M.R. wrote the behavioural section of the manuscript and J.B., V.S., W.C.W. and R.M.R. prepared the figures. All the authors discussed the results and commented on the manuscript.
Corresponding authors
Ethics declarations
Competing interests
A.L.H., J.B. and G.R.B. are shareholders in Ex Scientia Ltd, a University of Dundee spin-off company that has licensed the technology.
Supplementary information
Supplementary Information
This file contains Supplementary Figures 1-13, Supplementary Methods, Supplementary Tables 1-12 and Supplementary References – see contents for further details. (PDF 12182 kb)
Rights and permissions
About this article
Cite this article
Besnard, J., Ruda, G., Setola, V. et al. Automated design of ligands to polypharmacological profiles. Nature 492, 215–220 (2012). https://doi.org/10.1038/nature11691
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature11691
This article is cited by
-
A novel small molecule, AS1, reverses the negative hedonic valence of noxious stimuli
BMC Biology (2023)
-
Testing the limits of SMILES-based de novo molecular generation with curriculum and deep reinforcement learning
Nature Machine Intelligence (2023)
-
Identification of 5-HT2A receptor signaling pathways associated with psychedelic potential
Nature Communications (2023)
-
Chemogenetics for cell-type-specific modulation of signalling and neuronal activity
Nature Reviews Methods Primers (2023)
-
5-MeO-DMT: An atypical psychedelic with unique pharmacology, phenomenology & risk?
Psychopharmacology (2023)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.