Abstract
Brain-derived neurotrophic factor (BDNF) and its receptor TrkB are crucial for many forms of neuronal plasticity1,2,3,4,5,6, including structural long-term potentiation (sLTP)7,8, which is a correlate of an animal’s learning7,9,10,11,12. However, it is unknown whether BDNF release and TrkB activation occur during sLTP, and if so, when and where. Here, using a fluorescence resonance energy transfer-based sensor for TrkB and two-photon fluorescence lifetime imaging microscopy13,14,15,16, we monitor TrkB activity in single dendritic spines of CA1 pyramidal neurons in cultured murine hippocampal slices. In response to sLTP induction9,14,15,16, we find fast (onset < 1 min) and sustained (>20 min) activation of TrkB in the stimulated spine that depends on NMDAR (N-methyl-d-aspartate receptor) and CaMKII signalling and on postsynaptically synthesized BDNF. We confirm the presence of postsynaptic BDNF using electron microscopy to localize endogenous BDNF to dendrites and spines of hippocampal CA1 pyramidal neurons. Consistent with these findings, we also show rapid, glutamate-uncaging-evoked, time-locked BDNF release from single dendritic spines using BDNF fused to superecliptic pHluorin17,18,19. We demonstrate that this postsynaptic BDNF–TrkB signalling pathway is necessary for both structural and functional LTP20. Together, these findings reveal a spine-autonomous, autocrine signalling mechanism involving NMDAR–CaMKII-dependent BDNF release from stimulated dendritic spines and subsequent TrkB activation on these same spines that is crucial for structural and functional plasticity.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
References
Lohof, A. M., Ip, N. Y. & Poo, M. M. Potentiation of developing neuromuscular synapses by the neurotrophins NT-3 and BDNF. Nature 363, 350–353 (1993)
Kang, H., Welcher, A. A., Shelton, D. & Schuman, E. M. Neurotrophins and time: different roles for TrkB signaling in hippocampal long-term potentiation. Neuron 19, 653–664 (1997)
Minichiello, L. et al. Essential role for TrkB receptors in hippocampus-mediated learning. Neuron 24, 401–414 (1999)
Figurov, A., Pozzo-Miller, L. D., Olafsson, P., Wang, T. & Lu, B. Regulation of synaptic responses to high-frequency stimulation and LTP by neurotrophins in the hippocampus. Nature 381, 706–709 (1996)
Korte, M. et al. Hippocampal long-term potentiation is impaired in mice lacking brain-derived neurotrophic factor. Proc. Natl Acad. Sci. USA 92, 8856–8860 (1995)
Kovalchuk, Y., Hanse, E., Kafitz, K. W. & Konnerth, A. Postsynaptic induction of BDNF-mediated long-term potentiation. Science 295, 1729–1734 (2002)
Tanaka, J. et al. Protein synthesis and neurotrophin-dependent structural plasticity of single dendritic spines. Science 319, 1683–1687 (2008)
Lai, K.-O. O. et al. TrkB phosphorylation by Cdk5 is required for activity-dependent structural plasticity and spatial memory. Nat. Neurosci. 15, 1506–1515 (2012)
Matsuzaki, M., Honkura, N., Ellis-Davies, G. C. & Kasai, H. Structural basis of long-term potentiation in single dendritic spines. Nature 429, 761–766 (2004)
Okamoto, K., Nagai, T., Miyawaki, A. & Hayashi, Y. Rapid and persistent modulation of actin dynamics regulates postsynaptic reorganization underlying bidirectional plasticity. Nat. Neurosci. 7, 1104–1112 (2004)
Kim, I. H. et al. Disruption of Arp2/3 results in asymmetric structural plasticity of dendritic spines and progressive synaptic and behavioral abnormalities. J. Neurosci. 33, 6081–6092 (2013)
Kim, I. H., Wang, H., Soderling, S. H. & Yasuda, R. Loss of Cdc42 leads to defects in synaptic plasticity and remote memory recall. eLife 3, e02839 (2014)
Yasuda, R. Imaging spatiotemporal dynamics of neuronal signaling using fluorescence resonance energy transfer and fluorescence lifetime imaging microscopy. Curr. Opin. Neurobiol. 16, 551–561 (2006)
Harvey, C. D., Yasuda, R., Zhong, H. & Svoboda, K. The spread of Ras activity triggered by activation of a single dendritic spine. Science 321, 136–140 (2008)
Lee, S.-J. R. J., Escobedo-Lozoya, Y., Szatmari, E. M. & Yasuda, R. Activation of CaMKII in single dendritic spines during long-term potentiation. Nature 458, 299–304 (2009)
Murakoshi, H., Wang, H. & Yasuda, R. Local, persistent activation of Rho GTPases during plasticity of single dendritic spines. Nature 472, 100–104 (2011)
Miesenböck, G., De Angelis, D. A. & Rothman, J. E. Visualizing secretion and synaptic transmission with pH-sensitive green fluorescent proteins. Nature 394, 192–195 (1998)
Matsuda, N. et al. Differential activity-dependent secretion of brain-derived neurotrophic factor from axon and dendrite. J. Neurosci. 29, 14185–14198 (2009)
Dean, C. et al. Synaptotagmin-IV modulates synaptic function and long-term potentiation by regulating BDNF release. Nat. Neurosci. 12, 767–776 (2009)
Hedrick, N. G. et al. Rho GTPase complementation underlies BDNF-dependent homo- and heterosynaptic plasticity. Nature http://dx.doi.org/10.1038/nature19784 (2016)
Middlemas, D. S., Meisenhelder, J. & Hunter, T. Identification of TrkB autophosphorylation sites and evidence that phospholipase C-γ1 is a substrate of the TrkB receptor. J. Biol. Chem. 269, 5458–5466 (1994)
Vest, R. S., Davies, K. D., O’Leary, H., Port, J. D. & Bayer, K. U. Dual mechanism of a natural CaMKII inhibitor. Mol. Biol. Cell 18, 5024–5033 (2007)
Lu, W. et al. Subunit composition of synaptic AMPA receptors revealed by a single-cell genetic approach. Neuron 62, 254–268 (2009)
Huang, Y. Z., Pan, E., Xiong, Z.-Q. Q. & McNamara, J. O. Zinc-mediated transactivation of TrkB potentiates the hippocampal mossy fiber-CA3 pyramid synapse. Neuron 57, 546–558 (2008)
Dieni, S. et al. BDNF and its pro-peptide are stored in presynaptic dense core vesicles in brain neurons. J. Cell Biol. 196, 775–788 (2012)
Yang, J. et al. Neuronal release of proBDNF. Nat. Neurosci. 12, 113–115 (2009)
Lou, H. et al. Sorting and activity-dependent secretion of BDNF require interaction of a specific motif with the sorting receptor carboxypeptidase E. Neuron 45, 245–255 (2005)
Chen, X. et al. A chemical-genetic approach to studying neurotrophin signaling. Neuron 46, 13–21 (2005)
He, X.-P. et al. Conditional deletion of TrkB but not BDNF prevents epileptogenesis in the kindling model. Neuron 43, 31–42 (2004)
Luikart, B. W. et al. TrkB has a cell-autonomous role in the establishment of hippocampal Schaffer collateral synapses. J Neurosci. 25, 3774–3786 (2005)
Huang, Y. Z. & McNamara, J. O. Mutual regulation of Src family kinases and the neurotrophin receptor TrkB. J. Biol. Chem. 285, 8207–8217 (2010)
Zacharias, D. A., Violin, J. D., Newton, A. C. & Tsien, R. Y. Partitioning of lipid-modified monomeric GFPs into membrane microdomains of live cells. Science 296, 913–916 (2002)
Patterson, M. A., Szatmari, E. M. & Yasuda, R. AMPA receptors are exocytosed in stimulated spines and adjacent dendrites in a Ras-ERK-dependent manner during long-term potentiation. Proc. Natl Acad. Sci. USA 107, 15951–15956 (2010)
He, X.-P. P. et al. Conditional deletion of TrkB but not BDNF prevents epileptogenesis in the kindling model. Neuron 43, 31–42 (2004)
Xiong, Z. Q. & McNamara, J. O. Fleeting activation of ionotropic glutamate receptors sensitizes cortical neurons to complement attack. Neuron 36, 363–374 (2002)
Stoppini, L., Buchs, P. A. & Muller, D. A simple method for organotypic cultures of nervous tissue. J. Neurosci. Methods 37, 173–182 (1991)
Murakoshi, H., Lee, S.-J. J. & Yasuda, R. Highly sensitive and quantitative FRET-FLIM imaging in single dendritic spines using improved non-radiative YFP. Brain Cell Biol . 36, 31–42 (2008)
Pologruto, T. A., Sabatini, B. L. & Svoboda, K. ScanImage: flexible software for operating laser scanning microscopes. Biomed. Eng. Online 2, 13 (2003)
Pan, E. et al. Vesicular zinc promotes presynaptic and inhibits postsynaptic long-term potentiation of mossy fiber-CA3 synapse. Neuron 71, 1116–1126 (2011)
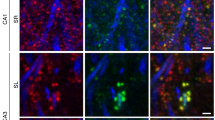
Milner, T. A., Waters, E. M., Robinson, D. C. & Pierce, J. P. in Neurodegeneration, Methods and Procedures (eds Manfredi, G. & Kawamata, H. ) 23–59 (Spring, 2011)
Peters, A., Palay, S. L. & Webster, H. D. The Fine Structure of the Nervous System 3rd edn (Oxford Univ. Press, 1991)
Acknowledgements
We thank A. West and Y. Huang for critical discussion. This work was supported by grants from the National Institutes of Health (F31NS078847 (S.C.H.), R01NS068410 (R.Y.), DP1NS096787 (R.Y.), R01NS05621 (J.O.M.), R01MH080047 (R.Y.), R01DA08259 (T.A.M.), R01HL098351 (T.A.M.), P01HL096571 (T.A.M.), and RO1NS030687 (B.L.H.)), the Wakeman Fellowship (S.C.H.), and Human Frontier Science Program (T.L.).
Author information
Authors and Affiliations
Contributions
S.C.H., N.G.H., R.Y. and J.O.M. designed experiments; S.C.H. and N.G.H. collected and analysed imaging data with assistance from C.E.H.; P.P.-B. and E.P. collected and analysed patch clamp data; T.A.M. collected electron microscopic images and analysed them with B.L.H.; T.L. performed in utero viral injections; S.C.H., N.G.H., R.Y. and J.O.M. analysed remaining data and wrote the paper. All authors discussed results and comments on this manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Additional information
Reviewer Information
Nature thanks B. Bingol, H. Zhang and the other anonymous reviewer(s) for their contribution to the peer review of this work.
Extended data figures and tables
Extended Data Figure 1 Design and development of a FRET-based sensor for TrkB activation.
a, Top, western blot analysis of cell extracts from HeLa cells stimulated with either BDNF or vehicle. Extracts were immunoprecipitated with an antibody for phosphorylated tyrosine residues (pTyr) and then probed with antibodies for TrkB and GFP. Bottom, immunoblot (IB) of BDNF and vehicle stimulated cell extracts before immunoprecipitation (IP) using antibodies for TrkB, GFP and actin. For source data, see Supplementary Fig. 1. b, FLIM images of TrkB and TrkBY816F activation acquired before and 2–6 min after BDNF stimulation (averaged multiple images taken over 5 min). Warmer colours indicate shorter lifetimes and higher TrkB activity. c, Time course of TrkB and TrkBY816F activation measured as the change in binding fraction of TrkB–eGFP or TrkBY816F–eGFP bound to mRFP1–PLC–mRFP1 before and after BDNF or vehicle stimulation. n = 22/8 TrkB plus BDNF, 9/4 TrkB plus vehicle, and 11/4 TrkBY816F plus BDNF (cells/experiments). d, TrkB activation (averaged over 6–10 min) for experiments in c. e, FLIM images of TrkB activation in a neuron in a mixed cortical dissociated culture before and after BDNF stimulation followed by K252a application at 30 min. f, Time course of TrkB activation measured as described in c before and after BDNF or NGF stimulation followed by K252a application. n = 8 BDNF and 4 NGF (neurons). g, TrkB activation (averaged over 10–30 min and 3–5 min following K252a application) for experiments in f. Data are mean ± s.e.m. *P < 0.05 as determined by a two-tailed unpaired samples t-test (g) or an analysis of variance (ANOVA) followed by Tukey’s method to correct for multiple comparisons. (d). **P < 0.05 as determined by a two-tailed paired samples t-test.
Extended Data Figure 2 Rescue of sLTP with TrkB–eGFP following postsynaptic TrkB knockout.
a, b, Time course (a) and quantification (b) of glutamate-uncaging-induced spine volume change for Trkbfl/fl hippocampal slices transfected with eGFP (Cre Neg), eGFP plus Cre (Cre Pos), and mCh, TrkB–eGFP and Cre (Cre Pos + TrkB–eGFP). n = 7/20 Cre Neg, 9/24 Cre Pos, and 5/11 Cre Pos + TrkB–eGFP (cells/spines). Data are mean ± s.e.m. *P < 0.05 as determined by an ANOVA followed by Tukey’s method to correct for multiple comparisons.
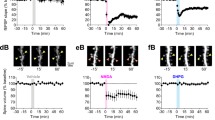
Extended Data Figure 3 Characterization of prolonged TrkB activation and spine volume change during single spine sLTP.
a, Prolonged time course of spine volume change after two-photon glutamate uncaging in rat hippocampal slices transfected with the TrkB sensor or eGFP. n = 50/54 for TrkB sensor (9/10 for experiments longer than 20 min) and 8/8 for eGFP (cells/spines). b, Prolonged time course of TrkB activation in stimulated spines, the base of the spine neck, adjacent spines, and the dendritic shaft adjacent to the stimulated spine. n = 50 cells with 54 stimulated spine, spine base, and dendrite plus 59 adjacent spine. c, d, Time course (c) and quantification (d) of the transient (averaged over 1–2 min) and sustained (averaged over 20–40 min) phases of glutamate uncaging-induced spine volume change in rat hippocampal slices in the absence and presence of anisomycin (25 μM). n = 12/14 control and 5/5 anisomycin (cells/spines). Data are mean ± s.e.m.
Extended Data Figure 4 Comparison of temporal dynamics of BDNF release, TrkB activation, and spine volume change during single spine sLTP.
a, Time course of normalized changes in TrkB activity and spine volume change (percentage of maximal activity and volume change). b, Magnified view of normalized changes of BDNF release, TrkB activation, and spine volume during and 1 min after the uncaging epoch.
Extended Data Figure 5 Determination of the specificity of glutamate uncaging evoked TrkB activation.
a, Time course of TrkB activation following glutamate uncaging before and at least 30 min after K252a application to the perfusion bath. n = 41/45 Ctrl and 4/9 K252a (cells/spines). b, Peak (averaged over 1–2 min) and sustained (averaged over 10–20 min) TrkB activation for experiments in a. c, Time course of spine volume change for experiments in a. d, Transient and sustained spine volume change for experiments in a. e–h, Similar experiments to a–d but in TrkbF616A hippocampal slices transfected with the TrkBF616A sensor before and at least 30 min after 1NMPP1 application (2 μM). n = 4/5 control and 3/6 1NMPP1 (cells/spines). i–l, Similar experiments to a–d but with the TrkB and TrkBY816F sensors. n = 9/10 control and 7/11 Y816F (cells/spines). Data are mean ± s.e.m. *P < 0.05 as determined by two-tailed unpaired samples t-test.
Extended Data Figure 6 Effect of temperature on the spatiotemporal dynamics of TrkB activation.
Time course of TrkB activation at room temperature (RT; 24–26 °C) and 30–32 °C in the stimulated and dendrite. n = 19/20 and 23/25 at 24–26 °C and 30–32 °C, respectively (spines/cells). Data are mean ± s.e.m.
Extended Data Figure 7 Effects of sensor expression levels on changes reported by the sensor.
a–d, Effect of TrkB–eGFP concentration as measured in individual neurons on corresponding change in binding fraction of the stimulated spine (a), change in spine volume (b), binding fraction before uncaging (basal binding fraction) (c), and change in binding fraction of the dendrite (d). n = 25/28 (cells/spines). Data are mean values and were fit to a linear regression model with corresponding coefficients of determination (R2) provided for each.
Extended Data Figure 8 Basal spine size and CaMKII activation in the presence and absence of post- synaptic BDNF.
a, b, Quantification (a) and representative two-photon images (f) of basal spine size/morphology in Bdnffl/fl slices transfected with eGFP or eGFP plus Cre (Cre Neg or Pos). n = 14/50 Cre Neg and 29/117 Cre Pos (cells/spines). Scale bar, 1 μm. c, d, Time course (c) and quantification (averaged over 0–45 s) (d) of CaMKII activation in Bdnffl/fl slices transfected with the CaMKII sensor or CaMKII plus Cre. n = 7/13 Cre Neg and 7/15 for Cre Pos (cells/spines). e, f, Time course and quantification of the transient phase of spine volume change for experiments in c. Data are mean ± s.e.m. *P < 0.05 as determined by a two-tailed unpaired samples t-test.
Extended Data Figure 9 Design and validation of BDNF–SEP.
a, Schematic of BDNF–SEP and BDNF–mRFP1. Pro, amino acids 19–128 of human BDNF; BDNF, amino acids 129–247 of human BDNF corresponding to the mature chain. b, Mechanistic model linking changes in SEP fluorescence with BDNF release. c, Change in BDNF–SEP fluorescence following glutamate uncaging under control, acidic (pH 6.5), and basic (pH 8.0) conditions. d, Confocal images of a CA1 pyramidal neuron transfected with eGFP and BDNF–mRFP1. Arrowheads indicate dendritic spines. e, Prolonged time course of BDNF–SEP fluorescence change (left) and spine volume change (right) in response to glutamate uncaging. n = 11/20 (cells/spines). f, g, Time course (f) and quantification (g) of spine volume change for experiments in Fig. 4c, d. n = 31/218 control, 6/82 TeTx, 2/29 POMC, 3/50 AP5, 2/46 AP5 + NBQX, 4/40 NBQX, and 7/88 CN21 (cells/spines). h, Data from Fig. 4d presented as median ± interquartile range. Data are mean ± s.e.m. unless otherwise indicated. *P < 0.05 as determined by an ANOVA followed by Dunnet’s method to correct for multiple comparisons. **P < 0.05 as determined by a Kruskal–Wallis test followed by Dunn’s test.
Extended Data Figure 10 CA1-LTP requires exogenous BDNF.
a, Time course of average EPSC amplitude changes recorded in CA1 pyramidal cells evoked by Schaffer collateral stimulation before and after LTP induction in the absence or presence of human-IgG or TrkB-Ig. Representative traces are above the graphs. n = 22 control, 9 HIgG, and 12 TrkB-Ig (animals). b, Quantification of EPSC amplitude changes averaged over 10–20 min following LTP induction. c, d, Time course (c) and quantification (d) of the transient and sustained glutamate-uncaging-induced spine volume change in rat hippocampal slices in the absence or presence of human-IgG or TrkB-Ig. n = 8/8 control, 6/8 TrkB-Ig, and 4/6 HIgG (cells/spines). e, Model of spine autonomous, autocrine, BDNF release and postsynaptic TrkB activation. Data are mean ± s.e.m. *P < 0.05 as determined by an ANOVA followed by Tukey’s method to correct for multiple comparisons
Supplementary information
Supplementary Notes
This file provides additional background and explanation for critical ideas and findings discussed in the main text. Additionally, it provides information regarding specific aspects of the Extended Data requiring further explanation. (PDF 272 kb)
Supplementary Figure
This file contains source data for the Western Blot data in Extended Data Figure 1. (PDF 98 kb)
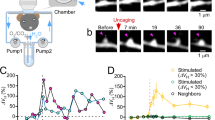
Glutamate-uncaging–evoked BDNF release from a dendritic spine
Two-photon video demonstrating the increase in BDNF-SEP fluorescence in response to the induction of sLTP by two-photon glutamate uncaging. Images were acquired at 8Hz. After baseline image acquisition (4 seconds/32 frames), an uncaging pulse was delivered at 0.5Hz (every 16 frames as indicated by arrow heads) for a total of 30 stimuli. The video was smoothed using a 3x3x2 3D Gaussian filter (ImageJ). Red fluorescence represents an mCherry-mCherry cell fill. Green fluorescence corresponds to BDNF-SEP. (AVI 2706 kb)
Rights and permissions
About this article
Cite this article
Harward, S., Hedrick, N., Hall, C. et al. Autocrine BDNF–TrkB signalling within a single dendritic spine. Nature 538, 99–103 (2016). https://doi.org/10.1038/nature19766
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature19766
This article is cited by
-
The role of neurotrophic factors in novel, rapid psychiatric treatments
Neuropsychopharmacology (2024)
-
Excitation–transcription coupling, neuronal gene expression and synaptic plasticity
Nature Reviews Neuroscience (2023)
-
Long-Lasting Impact of Sugar Intake on Neurotrophins and Neurotransmitters from Adolescence to Young Adulthood in Rat Frontal Cortex
Molecular Neurobiology (2023)
-
BDNF and Lactate as Modulators of Hippocampal CA3 Network Physiology
Cellular and Molecular Neurobiology (2023)
-
Role of Brain-Derived Neurotrophic Factor in Anxiety or Depression After Percutaneous Coronary Intervention
Molecular Neurobiology (2023)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.