Abstract
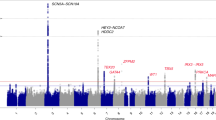
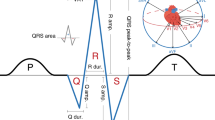
To identify genetic factors influencing cardiac conduction, we carried out a genome-wide association study of electrocardiographic time intervals in 6,543 Indian Asians. We identified association of a nonsynonymous SNP, rs6795970, in SCN10A (P = 2.8 × 10−15) with PR interval, a marker of cardiac atrioventricular conduction. Replication testing among 6,243 Indian Asians and 5,370 Europeans confirmed that rs6795970 (G>A) is associated with prolonged cardiac conduction (longer P-wave duration, PR interval and QRS duration, P = 10−5 to 10−20). SCN10A encodes NaV1.8, a sodium channel. We show that SCN10A is expressed in mouse and human heart tissue and that PR interval is shorter in Scn10a−/− mice than in wild-type mice. We also find that rs6795970 is associated with a higher risk of heart block (P < 0.05) and a lower risk of ventricular fibrillation (P = 0.01). Our findings provide new insight into the pathogenesis of cardiac conduction, heart block and ventricular fibrillation.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
References
Murray, C.J. & Lopez, A.D. Mortality by cause for eight regions of the world: Global Burden of Disease Study. Lancet 349, 1269–1276 (1997).
Lopez, A.D., Mathers, C.D., Ezzati, M., Jamison, D.T. & Murray, C.J. Global and regional burden of disease and risk factors, 2001: systematic analysis of population health data. Lancet 367, 1747–1757 (2006).
Huikuri, H.V., Castellanos, A. & Myerburg, R.J. Sudden death due to cardiac arrhythmias. N. Engl. J. Med. 345, 1473–1482 (2001).
Noseworthy, P.A. & Newton-Cheh, C. Genetic determinants of sudden cardiac death. Circulation 118, 1854–1863 (2008).
Knollmann, B.C. & Roden, D.M. A genetic framework for improving arrhythmia therapy. Nature 451, 929–936 (2008).
Schnabel, R.B. et al. Development of a risk score for atrial fibrillation (Framingham Heart Study): a community-based cohort study. Lancet 373, 739–745 (2009).
Zimetbaum, P.J. et al. Electrocardiographic predictors of arrhythmic death and total mortality in the multicenter unsustained tachycardia trial. Circulation 110, 766–769 (2004).
Vrtovec, B. et al. Prolonged QTc interval and high B-type natriuretic peptide levels together predict mortality in patients with advanced heart failure. Circulation 107, 1764–1769 (2003).
Schouten, E.G. et al. QT interval prolongation predicts cardiovascular mortality in an apparently healthy population. Circulation 84, 1516–1523 (1991).
Srinath Reddy, K., Shah, B., Varghese, C. & Ramadoss, A. Responding to the threat of chronic diseases in India. Lancet 366, 1744–1749 (2005).
Souslova, V.A., Fox, M., Wood, J.N. & Akopian, A.N. Cloning and characterization of a mouse sensory neuron tetrodotoxin-resistant voltage-gated sodium channel gene, Scn10a. Genomics 41, 201–209 (1997).
Akopian, A.N., Sivilotti, L. & Wood, J.N. A tetrodotoxin-resistant voltage-gated sodium channel expressed by sensory neurons. Nature 379, 257–262 (1996).
Zimmermann, K. et al. Sensory neuron sodium channel Nav1.8 is essential for pain at low temperatures. Nature 447, 855–858 (2007).
Djouhri, L. et al. The TTX-resistant sodium channel Nav1.8 (SNS/PN3): expression and correlation with membrane properties in rat nociceptive primary afferent neurons. J. Physiol. (Lond.) 550, 739–752 (2003).
Abrahamsen, B. et al. The cell and molecular basis of mechanical, cold and inflammatory pain. Science 321, 702–705 (2008).
Akopian, A.N. et al. The tetrodotoxin-resistant sodium channel SNS has a specialized function in pain pathways. Nat. Neurosci. 2, 541–548 (1999).
Renganathan, M., Cummins, T.R. & Waxman, S.G. Contribution of Na(v)1.8 sodium channels to action potential electrogenesis in DRG neurons. J. Neurophysiol. 86, 629–640 (2001).
Patrick Harty, T. & Waxman, S.G. Inactivation properties of sodium channel Nav1.8 maintain action potential amplitude in small DRG neurons in the context of depolarization. Mol. Pain 3, 12 (2007).
Eckel, R.H., Grundy, S.M. & Zimmet, P.Z. The metabolic syndrome. Lancet 365, 1415–1428 (2005).
Dekker, L.R. et al. Familial sudden death is an important risk factor for primary ventricular fibrillation: a case-control study in acute myocardial infarction patients. Circulation 114, 1140–1145 (2006).
Fitzgerald, E.M., Okuse, K., Wood, J.N., Dolphin, A.C. & Moss, S.J. cAMP-dependent phosphorylation of the tetrodotoxin-resistant voltage-dependent sodium channel SNS. J. Physiol. (Lond.) 516, 433–446 (1999).
Matsumoto, S. et al. Effect of 8-bromo-cAMP on the tetrodotoxin-resistant sodium (Nav 1.8) current in small-diameter nodose ganglion neurons. Neuropharmacology 52, 904–924 (2007).
Kerr, N.C., Holmes, F.E. & Wynick, D. Novel isoforms of the sodium channels Nav1.8 and Nav1.5 are produced by a conserved mechanism in mouse and rat. J. Biol. Chem. 279, 24826–24833 (2004).
Malik-Hall, M., Poon, W.Y., Baker, M.D., Wood, J.N. & Okuse, K. Sensory neuron proteins interact with the intracellular domains of sodium channel NaV1.8. Brain Res. Mol. Brain Res. 110, 298–304 (2003).
Tan, H.L. et al. A sodium-channel mutation causes isolated cardiac conduction disease. Nature 409, 1043–1047 (2001).
Lehnart, S.E. et al. Inherited arrhythmias: a National Heart, Lung, and Blood Institute and Office of Rare Diseases workshop consensus report about the diagnosis, phenotyping, molecular mechanisms, and therapeutic approaches for primary cardiomyopathies of gene mutations affecting ion channel function. Circulation 116, 2325–2345 (2007).
Pfeufer, A. et al. Common variants at ten loci modulate the QT interval duration in the QTSCD Study. Nat. Genet. 41, 407–414 (2009).
Lowe, J.K. et al. Genome-wide association studies in an isolated founder population from the Pacific Island of Kosrae. PLoS Genet. 5, e1000365 (2009).
Newton-Cheh, C. et al. Common variants at ten loci influence QT interval duration in the QTGEN Study. Nat. Genet. 41, 399–406 (2009).
Chambers, J.C. et al. Common genetic variation near MC4R is associated with waist circumference and insulin resistance. Nat. Genet. 40, 716–718 (2008).
Price, A.L. et al. Principal components analysis corrects for stratification in genome-wide association studies. Nat. Genet. 38, 904–909 (2006).
Pe'er, I., Yelensky, R., Altshuler, D. & Daly, M.J. Estimation of the multiple testing burden for genomewide association studies of nearly all common variants. Genet. Epidemiol. 32, 381–385 (2008).
Stagg, M.A. et al. Cytoskeletal protein 4.1R affects repolarization and regulates calcium handling in the heart. Circ. Res. 103, 855–863 (2008).
Acknowledgements
We thank the participants involved in the research. The LOLIPOP study was supported by the British Heart Foundation (SP/04/002) and by the Wellcome Trust (084723/Z/08/Z). Studies in the AGNES population were supported by the Netherlands Heart Foundation (Grant 2007B202) and the Leducq Foundation (Grant 05-CVD). J.Z. was supported by a BBSRC LOLA award (BB/F000227/1). R.K. was supported by the British Heart Foundation (Grant F/99089). M.N.W. is supported by the Biotechnology and Biological Sciences Research Council grant BB/F020481/1. W.M. is funded by the National Institute for Health Research Biomedical Research Centres scheme. J.N.W. is a member of the Wellcome Trust-funded London Pain Consortium. We thank G. Turner and M. Minett for maintaining the Scn10a-null mutant mouse colony, M.W. Tanck and R. Pazoki for help in statistical analyses and S. Rothery for technical assistance.
Author information
Authors and Affiliations
Contributions
Design and scientific rationale: J.C.C., J.Z., C.M.N.T., C.R.B., R.K., W.M., N.J.S., R.de.S., A.A.M.W., P.A., M.Y., J.S., P.E., J.N.W. and J.S.K. Obtained funding: J.C.C., C.M.N.T., C.R.B., R.K., N.J.S., A.A.M.W., P.A., M.Y., P.E., J.N.W. and J.S.K. Recruitment and characterization of participants: J.C.C., C.R.B., R.K., J.S.S., L.R.C.D., J.S.S.G.de.J., N.J.S., A.A.M.W., P.E. and J.S.K. Analyzed ECG readings: J.C.C., C.M.N.T., C.R.B., M.N., A.L., J.S.S., S.P., M.K.K., L.R.C.D., J.S.S.G.de.J. and A.A.M.W. Performed genotyping: J.C.C., C.R.B., W.Z., J.S.S., L.R.C.D., J.S.S.G.de.J., R.de.S., A.A.M.W., P.E. and J.S.K. PCR experiments: J.Z., C.M.N.T., R.K., U.S., I.E-H., N.J.S., J.C.C., J.N.W. and J.S.K. Mouse ECG studies: C.M.N.T., R.K., M.N., U.S., R.de.S., P.E., J.N.W., J.C.C. and J.S.K. Performed statistical analysis: J.C.C., C.M.N.T., C.R.B., W.Z., M.N., J.S.S., G.D., M.N.W., J.S.S.G.de.J., M.J.E.S., R.D.S., A.A.M.W., P.E. and J.S.K. Drafted manuscript: J.C.C., J.Z., C.M.N.T., J.S.S., R.de.S., J.S., P.E., J.N.W. and J.S.K. Critically revised manuscript: J.C.C., J.Z., C.M.N.T., C.R.B., W.Z., R.K., M.N., A.L., J.S.S., M.K.K., G.D., U.S., I.E-H., M.N.W., L.R.C.D., J.S.S.G.de.J., M.J.E.S., W.M., N.J.S., R.de.S., A.A.M.W., P.A., J.S., M.Y., P.E., J.N.W. and J.S.K.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Text and Figures
Supplementary Tables 1–7 and Supplementary Figures 1 and 2 (PDF 299 kb)
Rights and permissions
About this article
Cite this article
Chambers, J., Zhao, J., Terracciano, C. et al. Genetic variation in SCN10A influences cardiac conduction. Nat Genet 42, 149–152 (2010). https://doi.org/10.1038/ng.516
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ng.516
This article is cited by
-
Possible Antinociceptive Mechanisms Triggered by Nanomolar Ouabain Concentrations in Primary Sensory Neurons
Neuroscience and Behavioral Physiology (2021)
-
A novel loss-of-function mutation of PBK associated with human kidney stone disease
Scientific Reports (2020)
-
Inhibition of NaV1.8 prevents atrial arrhythmogenesis in human and mice
Basic Research in Cardiology (2020)
-
GAREM1 regulates the PR interval on electrocardiograms
Journal of Human Genetics (2018)
-
Loss-of-function mutations of SCN10A encoding NaV1.8 α subunit of voltage-gated sodium channel in patients with human kidney stone disease
Scientific Reports (2018)