Key Points
-
RING finger ubiquitin-protein ligases (E3s) are the most abundant class of E3 that mediate protein ubiquitylation (also known as ubiquitination). They regulate crucial cellular functions, such as the cell cycle, DNA repair, cell signalling and responses to hypoxia. Genetic alterations, including activating and inactivating mutations, gene amplifications, translocations and deletions, have been described for many RING finger E3s. RING finger E3s are validated oncogenes (such as MDM2) or tumour suppressor genes (such as BRCA1 and von Hippel–Lindau tumour suppressor (VHL)) because of their role in regulating crucial cell functions.
-
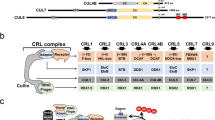
The cell cycle is regulated by the S phase kinase-associated protein 1 (SKP1)–cullin 1 (CUL1)–F-box protein (SCF) and anaphase-promoting complex/cyclosome (APC/C) multisubunit RING finger E3s. These complexes are targeted to specific substrates via interchangeable substrate recognition subunits, including F-box proteins for SCF and cell division cycle 20 (CDC20) and CDH1 for APC/C. These multisubunit E3s have a large number of substrates with oncogenic and tumour suppressive effects. Genetic alterations to components of these E3 complexes that result in loss of function (such as FBW7, CDH1 and CDC20) or gain of function (such as SKP2 and β-transducin repeat-containing protein (β-TrCP)) are implicated in the development of cancer.
-
RING finger E3s have central roles in DNA damage responses and DNA repair. For example, MDM2 targets p53 for degradation. MDM2 is amplified, overexpressed or activated in other ways in cancers and is a means of inactivating the tumour suppressor p53. The BRCA1 and the Fanconi anaemia (FANC) E3s have essential roles in the repair of DNA damage; both E3s function as tumour suppressors.
-
RING finger E3s have important roles in both positively and negatively regulating signal transduction. A prominent example of negative regulation is the CBL family of RING finger E3s that target activated receptor tyrosine kinases (RTKs) for degradation. Mutations that inactivate CBL E3 function have been described in myeloid neoplasms and result in the hyperactivation of RTKs and intracellular signalling pathways.
-
The response to hypoxia is regulated by the multisubunit CRL2VHL RING finger E3 and the single subunit RING finger E3 SIAH. The VHL complex targets the hypoxia-inducible factor-α (HIFα) transcription factors for proteasomal degradation, which prevents the expression of angiogenic and growth-promoting genes under normoxic conditions. Inactivating mutations of VHL are found in familial and sporadic clear cell cancer of the kidney, resulting in the stabilization of the HIFα transcription factor subunits and consequently abnormally high expression of angiogenic and growth genes. By contrast, the SIAH RING finger E3s stabilize HIFα under hypoxic conditions.
-
Targeting RING finger E3s for the treatment of cancer is being actively explored. For example, small-molecule inhibitors have been developed that interfere with the MDM2–p53 interaction or that inhibit MDM2 E3 activity, thus stabilizing p53. These approaches have demonstrated antitumour activity in preclinical studies, but the clinical efficacy of interfering with MDM2 function remains to be determined. Targeting the loss of activity of RING finger E3s that are tumour suppressors will require novel approaches such as the synthetic lethality that is induced by poly(ADP-ribose) polymerase (PARP) inhibition in cells that are deficient in BRCA1 or BRCA2.
Abstract
The ubiquitin-proteasome system has numerous crucial roles in physiology and pathophysiology. Fundamental to the specificity of this system are ubiquitin-protein ligases (E3s). Of these, the majority are RING finger and RING finger-related E3s. Many RING finger E3s have roles in processes that are central to the maintenance of genomic integrity and cellular homeostasis, such as the anaphase promoting complex/cyclosome (APC/C), the SKP1–cullin 1–F-box protein (SCF) E3s, MDM2, BRCA1, Fanconi anaemia proteins, CBL proteins, von Hippel–Lindau tumour suppressor (VHL) and SIAH proteins. As a result, many RING finger E3s are implicated in either the suppression or the progression of cancer. This Review summarizes current knowledge in this area.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
References
Lorick, K. L. et al. RING fingers mediate ubiquitin-conjugating enzyme (E2)-dependent ubiquitination. Proc. Natl Acad. Sci. USA 96, 11364–11369 (1999). This study determined that RING fingers are E2-binding ubiquitin ligase domains and that RING finger-containing proteins, including SIAH and BRCA1, have ubiquitin ligase activity.
Hanahan, D. & Weinberg, R. A. Hallmarks of cancer: the next generation. Cell 144, 646–674 (2011).
Nakayama, K. I. & Nakayama, K. Ubiquitin ligases: cell-cycle control and cancer. Nature Rev. Cancer 6, 369–381 (2006).
Skaar, J. R. & Pagano, M. Control of cell growth by the SCF and APC/C ubiquitin ligases. Curr. Opin. Cell Biol. 21, 816–824 (2009).
Frescas, D. & Pagano, M. Deregulated proteolysis by the F-box proteins SKP2 and β-TrCP: tipping the scales of cancer. Nature Rev. Cancer 8, 438–449 (2008).
Peters, J. M. The anaphase promoting complex/cyclosome: a machine designed to destroy. Nature Rev. Mol. Cell Biol. 7, 644–656 (2006). References 3–6 review the SCF and APC/C family of E3 ubiquitin ligases and their role in cell cycle control and cancer, and cite key primary literature.
Wang, Q. et al. Alterations of anaphase-promoting complex genes in human colon cancer cells. Oncogene 22, 1486–1490 (2003).
Wasch, R. & Engelbert, D. Anaphase-promoting complex-dependent proteolysis of cell cycle regulators and genomic instability of cancer cells. Oncogene 24, 1–10 (2005).
Garcia-Higuera, I. et al. Genomic stability and tumour suppression by the APC/C cofactor Cdh1. Nature Cell Biol. 10, 802–811 (2008).
Welcker, M. & Clurman, B. E. FBW7 ubiquitin ligase: a tumour suppressor at the crossroads of cell division, growth and differentiation. Nature Rev. Cancer 8, 83–93 (2008).
Guardavaccaro, D. et al. Control of chromosome stability by the β-TrCP-REST-Mad2 axis. Nature 452, 365–369 (2008).
Westbrook, T. F. et al. SCFβ-TRCP controls oncogenic transformation and neural differentiation through REST degradation. Nature 452, 370–374 (2008).
Wiggins, C. M. et al. BIM(EL), an intrinsically disordered protein, is degraded by 20S proteasomes in the absence of poly-ubiquitylation. J. Cell Sci. 124, 969–977 (2011).
Vousden, K. H. & Prives, C. Blinded by the light: the growing complexity of p53. Cell 137, 413–431 (2009).
Wade, M., Wang, Y. V. & Wahl, G. M. The p53 orchestra: Mdm2 and Mdmx set the tone. Trends Cell Biol. 20, 299–309 (2010).
Marine, J. C. & Lozano, G. Mdm2-mediated ubiquitylation: p53 and beyond. Cell Death Differ. 17, 93–102 (2010).
Lee, J. T. & Gu, W. The multiple levels of regulation by p53 ubiquitination. Cell Death Differ. 17, 86–92 (2010). References 14–17 review much of the relevant primary literature on p53, MDM2 and MDMX and their regulation.
Haupt, Y., Maya, R., Kazaz, A. & Oren, M. Mdm2 promotes the rapid degradation of p53. Nature 387, 296–299 (1997).
Kubbutat, M. H., Jones, S. N. & Vousden, K. H. Regulation of p53 stability by Mdm2. Nature 387, 299–303 (1997).
Honda, R., Tanaka, H. & Yasuda, H. Oncoprotein MDM2 is a ubiquitin ligase E3 for tumor suppressor p53. FEBS Lett. 420, 25–27 (1997).
Honda, R. & Yasuda, H. Activity of MDM2, a ubiquitin ligase, toward p53 or itself is dependent on the RING finger domain of the ligase. Oncogene 19, 1473–1476 (2000).
Fang, S., Jensen, J. P., Ludwig, R. L., Vousden, K. H. & Weissman, A. M. Mdm2 is a RING finger-dependent ubiquitin protein ligase for itself and p53. J. Biol. Chem. 275, 8945–8951 (2000). References 18–22 collectively establish that MDM2 is responsible for the degradation of p53 and is a RING finger-dependent E3 ubiquitin ligase for p53 and itself.
Jones, S. N., Roe, A. E., Donehower, L. A. & Bradley, A. Rescue of embryonic lethality in Mdm2-deficient mice by absence of p53. Nature 378, 206–208 (1995).
Montes de Oca Luna, R., Wagner, D. S. & Lozano, G. Rescue of early embryonic lethality in mdm2-deficient mice by deletion of p53. Nature 378, 203–206 (1995).
Mendrysa, S. M. et al. Tumor suppression and normal aging in mice with constitutively high p53 activity. Genes Dev. 20, 16–21 (2006).
Ringshausen, I., O'Shea, C. C., Finch, A. J., Swigart, L. B. & Evan, G. I. Mdm2 is critically and continuously required to suppress lethal p53 activity in vivo. Cancer Cell 10, 501–514 (2006). References 25 and 26 demonstrate the importance of the p53–MDM2 relationship in cancer and apoptosis using mouse models. They provide evidence supporting the concept that inducing moderate changes in p53 activity may be optimal in cancer therapies.
Uldrijan, S., Pannekoek, W. J. & Vousden, K. H. An essential function of the extreme C-terminus of MDM2 can be provided by MDMX. EMBO J. 26, 102–112 (2007).
Poyurovsky, M. V. et al. The Mdm2 RING domain C-terminus is required for supramolecular assembly and ubiquitin ligase activity. EMBO J. 26, 90–101 (2007).
Linke, K. et al. Structure of the MDM2/MDMX RING domain heterodimer reveals dimerization is required for their ubiquitylation in trans. Cell Death Differ. 15, 841–848 (2008). This paper elucidates the structure of the MDM2–MDMX RING finger dimer and provides important insights into function. The structure bears substantial similarity to the cIAP2 (also known as BIRC3) dimer, which was also solved by the Day laboratory.
Tanimura, S. et al. MDM2 interacts with MDMX through their RING finger domains. FEBS Lett. 447, 5–9 (1999).
Okamoto, K., Taya, Y. & Nakagama, H. Mdmx enhances p53 ubiquitination by altering the substrate preference of the Mdm2 ubiquitin ligase. FEBS Lett. 583, 2710–2714 (2009).
Linares, L. K., Hengstermann, A., Ciechanover, A., Muller, S. & Scheffner, M. HdmX stimulates Hdm2-mediated ubiquitination and degradation of p53. Proc. Natl Acad. Sci. USA 100, 12009–12014 (2003).
Stad, R. et al. Mdmx stabilizes p53 and Mdm2 via two distinct mechanisms. EMBO Rep. 2, 1029–1034 (2001).
Pomerantz, J. et al. The Ink4a tumor suppressor gene product, p19Arf, interacts with MDM2 and neutralizes MDM2's inhibition of p53. Cell 92, 713–723 (1998).
Weber, J. D., Taylor, L. J., Roussel, M. F., Sherr, C. J. & Bar-Sagi, D. Nucleolar Arf sequesters Mdm2 and activates p53. Nature Cell Biol. 1, 20–26 (1999).
Saporita, A. J., Maggi, L. B. J., Apicelli, A. J. & Weber, J. D. Therapeutic targets in the ARF tumor suppressor pathway. Curr. Med. Chem. 14, 1815–1827 (2007).
Zhang, Y. & Lu, H. Signaling to p53: ribosomal proteins find their way. Cancer Cell 16, 369–377 (2009).
Horn, H. F. & Vousden, K. H. Cooperation between the ribosomal proteins L5 and L11 in the p53 pathway. Oncogene 27, 5774–5784 (2008).
Dai, M. S. & Lu, H. Inhibition of MDM2-mediated p53 ubiquitination and degradation by ribosomal protein L5. J. Biol. Chem. 279, 44475–44482 (2004).
Lohrum, M. A., Ashcroft, M., Kubbutat, M. H. & Vousden, K. H. Identification of a cryptic nucleolar-localization signal in MDM2. Nature Cell Biol. 2, 179–181 (2000).
Issaeva, N. et al. Small molecule RITA binds to p53, blocks p53-HDM-2 interaction and activates p53 function in tumors. Nature Med. 10, 1321–1328 (2004). This paper demonstrates that binding a small molecule to p53 can inhibit its degradation and can activate a p53 response in tumours.
Zhao, C. Y., Szekely, L., Bao, W. & Selivanova, G. Rescue of p53 function by small-molecule RITA in cervical carcinoma by blocking E6-mediated degradation. Cancer Res. 70, 3372–3381 (2010).
Vassilev, L. T. et al. In vivo activation of the p53 pathway by small-molecule antagonists of MDM2. Science 303, 844–848 (2004). This paper demonstrates that a small molecule that blocks the p53-binding site on MDM2 can activate a p53 response in tumours.
Patterson, D. M. et al. Effect of MDM2 and vascular endothelial growth factor inhibition on tumor angiogenesis and metastasis in neuroblastoma. Angiogenesis 12 Apr 2011 (doi:10.1007/s10456-011-9210-8).
Tovar, C. et al. MDM2 antagonists boost antitumor effect of androgen withdrawal: implications for therapy of prostate cancer. Mol. Cancer 10, 49 (2011).
Bernal, F., Tyler, A. F., Korsmeyer, S. J., Walensky, L. D. & Verdine, G. L. Reactivation of the p53 tumor suppressor pathway by a stapled p53 peptide. J. Am. Chem. Soc. 129, 2456–2457 (2007).
Lai, Z. et al. Differentiation of Hdm2-mediated p53 ubiquitination and Hdm2 autoubiquitination activity by small molecular weight inhibitors. Proc. Natl Acad. Sci. USA 99, 14734–14739 (2002).
Yang, Y. et al. Small molecule inhibitors of HDM2 ubiquitin ligase activity stabilize and activate p53 in cells. Cancer Cell 7, 547–559 (2005). This paper provides proof-of-principle that blocking the ubiquitin ligase activity of MDM2 can activate a p53 response in cells and can kill tumour cells.
Kitagaki, J., Agama, K. K., Pommier, Y., Yang, Y. & Weissman, A. M. Targeting tumor cells expressing p53 with a water-soluble inhibitor of Hdm2. Mol. Cancer Ther. 7, 2445–2454 (2008).
Welcsh, P. L. & King, M. C. BRCA1 and BRCA2 and the genetics of breast and ovarian cancer. Hum. Mol. Genet. 10, 705–713 (2001).
Scully, R. et al. Dynamic changes of BRCA1 subnuclear location and phosphorylation state are initiated by DNA damage. Cell 90, 425–435 (1997).
Scully, R. et al. Association of BRCA1 with Rad51 in mitotic and meiotic cells. Cell 88, 265–275 (1997).
Moynahan, M. E., Chiu, J. W., Koller, B. H. & Jasin, M. Brca1 controls homology-directed DNA repair. Mol. Cell 4, 511–518 (1999).
Snouwaert, J. N. et al. BRCA1 deficient embryonic stem cells display a decreased homologous recombination frequency and an increased frequency of non-homologous recombination that is corrected by expression of a brca1 transgene. Oncogene 18, 7900–7907 (1999).
Huen, M. S., Sy, S. M. & Chen, J. BRCA1 and its toolbox for the maintenance of genome integrity. Nature Rev. Mol. Cell Biol. 11, 138–148 (2010). This review summarizes the role of BRCA1 in DNA repair.
Hashizume, R. et al. The RING heterodimer BRCA1-BARD1 is a ubiquitin ligase inactivated by a breast cancer-derived mutation. J. Biol. Chem. 276, 14537–14540 (2001). This paper establishes the importance of the BRCA1–BARD1 dimer in ubiquitin ligase function and demonstrates that a disease-associated mutation in the BRCA1 RING finger results in loss of activity.
Xia, Y., Pao, G. M., Chen, H. W., Verma, I. M. & Hunter, T. Enhancement of BRCA1 E3 ubiquitin ligase activity through direct interaction with the BARD1 protein. J. Biol. Chem. 278, 5255–5263 (2003).
Joukov, V., Chen, J., Fox, E. A., Green, J. B. & Livingston, D. M. Functional communication between endogenous BRCA1 and its partner, BARD1, during Xenopus laevis development. Proc. Natl Acad. Sci. USA 98, 12078–12083 (2001).
Meza, J. E., Brzovic, P. S., King, M. C. & Klevit, R. E. Mapping the functional domains of BRCA1. Interaction of the ring finger domains of BRCA1 and BARD1. J. Biol. Chem. 274, 5659–5665 (1999).
Brzovic, P. S., Rajagopal, P., Hoyt, D. W., King, M. C. & Klevit, R. E. Structure of a BRCA1-BARD1 heterodimeric RING-RING complex. Nature Struct. Biol. 8, 833–837 (2001). This paper elucidates the structure of the BRCA1–BARD1 heterodimer by NMR. This is the first structure of a RING finger dimer.
Morris, J. R. et al. The SUMO modification pathway is involved in the BRCA1 response to genotoxic stress. Nature 462, 886–890 (2009).
Yu, X., Fu, S., Lai, M., Baer, R. & Chen, J. BRCA1 ubiquitinates its phosphorylation-dependent binding partner CtIP. Genes Dev. 20, 1721–1726 (2006).
Barber, L. J. & Boulton, S. J. BRCA1 ubiquitylation of CtIP: just the tIP of the iceberg? DNA Repair 5, 1499–1504 (2006).
Ma, Y. et al. The breast cancer susceptibility gene BRCA1 regulates progesterone receptor signaling in mammary epithelial cells. Mol. Endocrinol. 20, 14–34 (2006).
Fan, S. et al. Role of direct interaction in BRCA1 inhibition of estrogen receptor activity. Oncogene 20, 77–87 (2001).
Eakin, C. M., Maccoss, M. J., Finney, G. L. & Klevit, R. E. Estrogen receptor α is a putative substrate for the BRCA1 ubiquitin ligase. Proc. Natl Acad. Sci. USA 104, 5794–5799 (2007).
Poole, A. J. et al. Prevention of Brca1-mediated mammary tumorigenesis in mice by a progesterone antagonist. Science 314, 1467–1470 (2006).
Katiyar, P., Ma, Y., Riegel, A., Fan, S. & Rosen, E. M. Mechanism of BRCA1-mediated inhibition of progesterone receptor transcriptional activity. Mol. Endocrinol. 23, 1135–1146 (2009).
Ma, Y. et al. BRCA1 regulates acetylation and ubiquitination of estrogen receptor-α. Mol. Endocrinol. 24, 76–90 (2010).
Li, W., Xiao, C., Vonderhaar, B. K. & Deng, C. X. A role of estrogen/ERα signaling in BRCA1-associated tissue-specific tumor formation. Oncogene 26, 7204–7212 (2007).
Jones, L. P. et al. Activation of estrogen signaling pathways collaborates with loss of Brca1 to promote development of ERα-negative and ERα-positive mammary preneoplasia and cancer. Oncogene 27, 794–802 (2008).
Eisen, A. et al. Breast cancer risk following bilateral oophorectomy in BRCA1 and BRCA2 mutation carriers: an international case-control study. J. Clin. Oncol. 23, 7491–7496 (2005).
Moldovan, G. L. & D'Andrea, A. D. How the fanconi anemia pathway guards the genome. Annu. Rev. Genet. 43, 223–249 (2009). This review summarizes a large body of work on the FANC ubiquitin ligase and its function.
Vaz, F. et al. Mutation of the RAD51C gene in a Fanconi anemia-like disorder. Nature Genet. 42, 406–409 (2010).
Stoepker, C. et al. SLX4, a coordinator of structure-specific endonucleases, is mutated in a new Fanconi anemia subtype. Nature Genet. 43, 138–141 (2011).
Machida, Y. J. et al. UBE2T is the E2 in the Fanconi anemia pathway and undergoes negative autoregulation. Mol. Cell 23, 589–596 (2006).
Eletr, Z. M., Huang, D. T., Duda, D. M., Schulman, B. A. & Kuhlman, B. E2 conjugating enzymes must disengage from their E1 enzymes before E3-dependent ubiquitin and ubiquitin-like transfer. Nature Struct. Mol. Biol. 12, 933–934 (2005).
Knipscheer, P. et al. The Fanconi anemia pathway promotes replication-dependent DNA interstrand cross-link repair. Science 326, 1698–1701 (2009).
MacKay, C. et al. Identification of KIAA1018/FAN1, a DNA repair nuclease recruited to DNA damage by monoubiquitinated FANCD2. Cell 142, 65–76 (2010).
Smogorzewska, A. et al. A genetic screen identifies FAN1, a Fanconi anemia-associated nuclease necessary for DNA interstrand crosslink repair. Mol. Cell 39, 36–47 (2010).
Kratz, K. et al. Deficiency of FANCD2-associated nuclease KIAA1018/FAN1 sensitizes cells to interstrand crosslinking agents. Cell 142, 77–88 (2010).
Kee, Y., Kim, J. M. & D'Andrea, A. D. Regulated degradation of FANCM in the Fanconi anemia pathway during mitosis. Genes Dev. 23, 555–560 (2009).
Farmer, H. et al. Targeting the DNA repair defect in BRCA mutant cells as a therapeutic strategy. Nature 434, 917–921 (2005).
Bryant, H. E. et al. Specific killing of BRCA2-deficient tumours with inhibitors of poly(ADP-ribose) polymerase. Nature 434, 913–917 (2005). References 83 and 84 demonstrate the synthetic lethality of PARP inhibition in cells deficient in BRCA1 or BRCA2, and are the basis of clinical trials using PARP inhibitors in patients with BRCA1 and BRCA2 mutations.
Fong, P. C. et al. Inhibition of poly(ADP-ribose) polymerase in tumors from BRCA mutation carriers. N. Engl. J. Med. 361, 123–134 (2009).
Audeh, M. W. et al. Oral poly(ADP-ribose) polymerase inhibitor olaparib in patients with BRCA1 or BRCA2 mutations and recurrent ovarian cancer: a proof-of-concept trial. Lancet 376, 245–251 (2010).
Tutt, A. et al. Oral poly(ADP-ribose) polymerase inhibitor olaparib in patients with BRCA1 or BRCA2 mutations and advanced breast cancer: a proof-of-concept trial. Lancet 376, 235–244 (2010).
Fujita, Y. et al. Hakai, a c-Cbl-like protein, ubiquitinates and induces endocytosis of the E-cadherin complex. Nature Cell Biol. 4, 222–231 (2002).
Qiu, X. B. & Goldberg, A. L. Nrdp1/FLRF is a ubiquitin ligase promoting ubiquitination and degradation of the epidermal growth factor receptor family member, ErbB3. Proc. Natl Acad. Sci. USA 99, 14843–14848 (2002).
Xu, W. et al. Chaperone-dependent E3 ubiquitin ligase CHIP mediates a degradative pathway for c-ErbB2/Neu. Proc. Natl Acad. Sci. USA 99, 12847–12852 (2002).
Zhou, P. et al. ErbB2 degradation mediated by the co-chaperone protein CHIP. J. Biol. Chem. 278, 13829–13837 (2003).
Li, Y., Kumar, K. G., Tang, W., Spiegelman, V. S. & Fuchs, S. Y. Negative regulation of prolactin receptor stability and signaling mediated by SCFβ-TrCP E3 ubiquitin ligase. Mol. Cell. Biol. 24, 4038–4048 (2004).
van Kerkhof, P., Putters, J. & Strous, G. J. The ubiquitin ligase SCFβTrCP regulates the degradation of the growth hormone receptor. J. Biol. Chem. 282, 20475–20483 (2007).
Clevers, H. Wnt/β-catenin signaling in development and disease. Cell 127, 469–480 (2006).
Staudt, L. M. Oncogenic activation of NF-κB. Cold Spring Harb. Perspect. Biol. 2, a000109 (2010).
Blume-Jensen, P. & Hunter, T. Oncogenic kinase signalling. Nature 411, 355–365 (2001).
Acconcia, F., Sigismund, S. & Polo, S. Ubiquitin in trafficking: the network at work. Exp. Cell Res. 315, 1610–1618 (2009).
Ettenberg, S. A. et al. cbl-b inhibits EGF-receptor-induced apoptosis by enhancing ubiquitination and degradation of activated receptors. Mol. Cell Biol. Res. Commun. 2, 111–118 (1999).
Levkowitz, G. et al. c-Cbl/Sli-1 regulates endocytic sorting and ubiquitination of the epidermal growth factor receptor. Genes Dev. 12, 3663–3674 (1998).
Miyake, S., Mullane-Robinson, K. P., Lill, N. L., Douillard, P. & Band, H. Cbl-mediated negative regulation of platelet-derived growth factor receptor-dependent cell proliferation. A critical role for Cbl tyrosine kinase-binding domain. J. Biol. Chem. 274, 16619–16628 (1999).
Joazeiro, C. A. et al. The tyrosine kinase negative regulator c-Cbl as a RING-type, E2-dependent ubiquitin-protein ligase. Science 286, 309–312 (1999).
Levkowitz, G. et al. Ubiquitin ligase activity and tyrosine phosphorylation underlie suppression of growth factor signaling by c-Cbl/Sli-1. Mol. Cell 4, 1029–1040 (1999).
Yokouchi, M. et al. Ligand-induced ubiquitination of the epidermal growth factor receptor involves the interaction of the c-Cbl RING finger and UbcH7. J. Biol. Chem. 274, 31707–31712 (1999). References 100–103 demonstrate that CBL proteins are RING finger ubiquitin ligases that ubiquitylate and target activated RTKs for degradation.
Nau, M. M. & Lipkowitz, S. in Cbl Proteins (ed. Tsygankov, A.) 3–25 (Nova Science Publishers, New York, 2008).
Zheng, N., Wang, P., Jeffrey, P. D. & Pavletich, N. P. Structure of a c-Cbl-UbcH7 complex: RING domain function in ubiquitin-protein ligases. Cell 102, 533–539 (2000). This paper describes the first crystal structure of a RING finger–E2 complex. The sites of RING finger–E2 interaction have held up through subsequent structure and structure–function analyses.
Kassenbrock, C. K. & Anderson, S. M. Regulation of ubiquitin protein ligase activity in c-Cbl by phosphorylation-induced conformational change and constitutive activation by tyrosine to glutamate point mutations. J. Biol. Chem. 279, 28017–28027 (2004).
Ryan, P. E., Sivadasan-Nair, N., Nau, M. M., Nicholas, S. & Lipkowitz, S. The N-terminus of Cbl-c regulates ubiquitin ligase activity by modulating affinity for the ubiquitin conjugating enzyme. J. Biol. Chem. 285, 23687–23698 (2010).
Huang, F., Kirkpatrick, D., Jiang, X., Gygi, S. & Sorkin, A. Differential regulation of EGF receptor internalization and degradation by multiubiquitination within the kinase domain. Mol. Cell 21, 737–748 (2006).
Mosesson, Y. et al. Endocytosis of receptor tyrosine kinases is driven by monoubiquitylation, not polyubiquitylation. J. Biol. Chem. 278, 21323–21326 (2003).
Schmidt, M. H. & Dikic, I. The Cbl interactome and its functions. Nature Rev. Mol. Cell Biol. 6, 907–918 (2005).
Langdon, W. Y., Hartley, J. W., Klinken, S. P., Ruscetti, S. K. & Morse, H. C. v-cbl, an oncogene from a dual-recombinant murine retrovirus that induces early B-lineage lymphomas. Proc. Natl Acad. Sci. USA 86, 1168–1172 (1989).
Bonita, D. P., Miyake, S., Lupher, M. L. Jr, Langdon, W. Y. & Band, H. Phosphotyrosine binding domain-dependent upregulation of the platelet-derived growth factor receptor α signaling cascade by transforming mutants of Cbl: implications for Cbl's function and oncogenicity. Mol. Cell. Biol. 17, 4597–4610 (1997).
Kales, S. C., Ryan, P. E., Nau, M. M. & Lipkowitz, S. Cbl and human myeloid neoplasms: the Cbl oncogene comes of age. Cancer Res. 70, 4789–4794 (2010).
Naramura, M., Nandwani, N., Gu, H., Band, V. & Band, H. Rapidly fatal myeloproliferative disorders in mice with deletion of Casitas B-cell lymphoma (Cbl) and Cbl-b in hematopoietic stem cells. Proc. Natl Acad. Sci. USA 107, 16274–16279 (2010).
Reindl, C. et al. CBL exon 8/9 mutants activate the FLT3 pathway and cluster in core binding factor/11q deletion acute myeloid leukemia/myelodysplastic syndrome subtypes. Clin. Cancer Res. 15, 2238–2247 (2009).
Sanada, M. et al. Gain-of-function of mutated C-CBL tumour suppressor in myeloid neoplasms. Nature 460, 904–908 (2009).
Sargin, B. et al. Flt3-dependent transformation by inactivating c-Cbl mutations in AML. Blood 110, 1004–1012 (2007).
Gilliland, D. G. & Griffin, J. D. Role of FLT3 in leukemia. Curr. Opin. Hematol. 9, 274–281 (2002).
Grand, F. H. et al. Frequent CBL mutations associated with 11q acquired uniparental disomy in myeloproliferative neoplasms. Blood 113, 6182–6192 (2009).
Rathinam, C., Thien, C. B., Flavell, R. A. & Langdon, W. Y. Myeloid leukemia development in c-Cbl RING finger mutant mice is dependent on FLT3 signaling. Cancer Cell 18, 341–352 (2010).
Tan, Y. H. et al. CBL is frequently altered in lung cancers: its relationship to mutations in MET and EGFR tyrosine kinases. PLoS ONE 5, e8972 (2010).
Peschard, P. & Park, M. Escape from Cbl-mediated downregulation: a recurrent theme for oncogenic deregulation of receptor tyrosine kinases. Cancer Cell 3, 519–523 (2003).
Ryan, P. E., Davies, G. C., Nau, M. M. & Lipkowitz, S. Regulating the regulator: negative regulation of Cbl ubiquitin ligases. Trends Biochem. Sci. 31, 79–88 (2006).
Thien, C. B. et al. c-Cbl promotes T cell receptor-induced thymocyte apoptosis by activating the phosphatidylinositol 3-kinase/Akt pathway. J. Biol. Chem. 285, 10969–10981 (2010).
Bachmaier, K. et al. Negative regulation of lymphocyte activation and autoimmunity by the molecular adaptor Cbl-b. Nature 403, 211–216 (2000).
Chiang, Y. J. et al. Cbl-b regulates the CD28 dependence of T-cell activation. Nature 403, 216–220 (2000).
Jeon, M. S. et al. Essential role of the E3 ubiquitin ligase Cbl-b in T cell anergy induction. Immunity 21, 167–177 (2004).
Chiang, J. Y., Jang, I. K., Hodes, R. & Gu, H. Ablation of Cbl-b provides protection against transplanted and spontaneous tumors. J. Clin. Invest. 117, 1029–1036 (2007).
Loeser, S. et al. Spontaneous tumor rejection by cbl-b-deficient CD8+ T cells. J. Exp. Med. 204, 879–891 (2007).
Paolino, M. et al. Essential role of E3 ubiquitin ligase activity in Cbl-b-regulated T cell functions. J. Immunol. 186, 2138–2147 (2011).
Latif, F. et al. Identification of the von Hippel-Lindau disease tumor suppressor gene. Science 260, 1317–1320 (1993).
Gnarra, J. R. et al. Mutations of the VHL tumour suppressor gene in renal carcinoma. Nature Genet. 7, 85–90 (1994).
Herman, J. G., Graff, J. R., Myohanen, S., Nelkin, B. D. & Baylin, S. B. Methylation-specific PCR: a novel PCR assay for methylation status of CpG islands. Proc. Natl Acad. Sci. USA 93, 9821–9826 (1996).
Iliopoulos, O., Kibel, A., Gray, S. & Kaelin, W. G. Jr. Tumour suppression by the human von Hippel-Lindau gene product. Nature Med. 1, 822–826 (1995).
Kamura, T. et al. Rbx1, a component of the VHL tumor suppressor complex and SCF ubiquitin ligase. Science 284, 657–661 (1999). This paper demonstrates the importance of the small RING finger protein RBX1 in the activity of the CRL2VHL E3 ubiquitin ligase. Contemporaneous papers from the Deshaies, Elledge, Harper, Pan and Xiong laboratories established the importance of this protein for the SCF family of CRL E3s.
Kibel, A., Iliopoulos, O., DeCaprio, J. A. & Kaelin, W. G. Jr. Binding of the von Hippel-Lindau tumor suppressor protein to Elongin B and C. Science 269, 1444–1446 (1995).
Pause, A. et al. The von Hippel-Lindau tumor-suppressor gene product forms a stable complex with human CUL-2, a member of the Cdc53 family of proteins. Proc. Natl Acad. Sci. USA 94, 2156–2161 (1997).
Iwai, K. et al. Identification of the von Hippel-lindau tumor-suppressor protein as part of an active E3 ubiquitin ligase complex. Proc. Natl Acad. Sci. USA 96, 12436–12441 (1999).
Lisztwan, J., Imbert, G., Wirbelauer, C., Gstaiger, M. & Krek, W. The von Hippel-Lindau tumor suppressor protein is a component of an E3 ubiquitin-protein ligase activity. Genes Dev. 13, 1822–1833 (1999). References 138 and 139 demonstrate that the VHL complex is an E3 ubiquitin ligase and that disease-associated mutations of the VHL protein abrogate E3 ubiquitin ligase activity.
Maxwell, P. H. et al. The tumour suppressor protein VHL targets hypoxia-inducible factors for oxygen-dependent proteolysis. Nature 399, 271–275 (1999).
Ohh, M. et al. Ubiquitination of hypoxia-inducible factor requires direct binding to the β-domain of the von Hippel-Lindau protein. Nature Cell Biol. 2, 423–427 (2000).
Gnarra, J. R. et al. Post-transcriptional regulation of vascular endothelial growth factor mRNA by the product of the VHL tumor suppressor gene. Proc. Natl Acad. Sci. USA 93, 10589–10594 (1996).
Iliopoulos, O., Levy, A. P., Jiang, C., Kaelin, W. G. Jr & Goldberg, M. A. Negative regulation of hypoxia-inducible genes by the von Hippel-Lindau protein. Proc. Natl Acad. Sci. USA 93, 10595–10599 (1996).
Ivan, M. et al. HIFα targeted for VHL-mediated destruction by proline hydroxylation: implications for O2 sensing. Science 292, 464–468 (2001).
Jaakkola, P. et al. Targeting of HIF-α to the von Hippel-Lindau ubiquitylation complex by O2-regulated prolyl hydroxylation. Science 292, 468–472 (2001).
Hon, W. C. et al. Structural basis for the recognition of hydroxyproline in HIF-1 α by pVHL. Nature 417, 975–978 (2002).
Epstein, A. C. et al. C. elegans EGL-9 and mammalian homologs define a family of dioxygenases that regulate HIF by prolyl hydroxylation. Cell 107, 43–54 (2001).
Kondo, K., Kim, W. Y., Lechpammer, M. & Kaelin, W. G. Jr. Inhibition of HIF2α is sufficient to suppress pVHL-defective tumor growth. PLoS Biol. 1, e83 (2003).
Kondo, K., Klco, J., Nakamura, E., Lechpammer, M. & Kaelin, W. G. Jr. Inhibition of HIF is necessary for tumor suppression by the von Hippel-Lindau protein. Cancer Cell 1, 237–246 (2002).
Maranchie, J. K. et al. The contribution of VHL substrate binding and HIF1-α to the phenotype of VHL loss in renal cell carcinoma. Cancer Cell 1, 247–255 (2002).
Zimmer, M., Doucette, D., Siddiqui, N. & Iliopoulos, O. Inhibition of hypoxia-inducible factor is sufficient for growth suppression of VHL−/− tumors. Mol. Cancer Res. 2, 89–95 (2004).
Nakayama, K. et al. Siah2 regulates stability of prolyl-hydroxylases, controls HIF1α abundance, and modulates physiological responses to hypoxia. Cell 117, 941–952 (2004).
House, C. M., Moller, A. & Bowtell, D. D. Siah proteins: novel drug targets in the Ras and hypoxia pathways. Cancer Res. 69, 8835–8838 (2009).
Crawford, L. J., Walker, B. & Irvine, A. E. Proteasome inhibitors in cancer therapy. J. Cell Commun. Signal. 5, 101–110 (2011).
Dominguez, C. et al. Structural model of the UbcH5B/CNOT4 complex revealed by combining NMR, mutagenesis, and docking approaches. Structure 12, 633–644 (2004).
Linehan, W. M., Srinivasan, R. & Schmidt, L. S. The genetic basis of kidney cancer: a metabolic disease. Nature Rev. Urol. 7, 277–285 (2010).
Isaacs, J. S. et al. HIF overexpression correlates with biallelic loss of fumarate hydratase in renal cancer: novel role of fumarate in regulation of HIF stability. Cancer Cell 8, 143–153 (2005).
Vitari, A. C. et al. COP1 is a tumour suppressor that causes degradation of ETS transcription factors. Nature 474, 403–406 (2011).
Migliorini, D. et al. Cop1 constitutively regulates c-Jun protein stability and functions as a tumor suppressor in mice. J. Clin. Invest. 121, 1329–1343 (2011).
Hsu, J. Y., Reimann, J. D., Sorensen, C. S., Lukas, J. & Jackson, P. K. E2F-dependent accumulation of hEmi1 regulates S phase entry by inhibiting APCCdh1. Nature Cell Biol. 4, 358–366 (2002).
Lehman, N. L., Verschuren, E. W., Hsu, J. Y., Cherry, A. M. & Jackson, P. K. Overexpression of the anaphase promoting complex/cyclosome inhibitor Emi1 leads to tetraploidy and genomic instability of p53-deficient cells. Cell Cycle 5, 1569–1573 (2006).
Reimann, J. D. et al. Emi1 is a mitotic regulator that interacts with Cdc20 and inhibits the anaphase promoting complex. Cell 105, 645–655 (2001).
O'Neil, J. et al. FBW7 mutations in leukemic cells mediate NOTCH pathway activation and resistance to γ-secretase inhibitors. J. Exp. Med. 204, 1813–1824 (2007).
Bloom, J. & Pagano, M. Deregulated degradation of the cdk inhibitor p27 and malignant transformation. Semin. Cancer Biol. 13, 41–47 (2003).
Gstaiger, M. et al. Skp2 is oncogenic and overexpressed in human cancers. Proc. Natl Acad. Sci. USA 98, 5043–5048 (2001).
Latres, E. et al. Role of the F-box protein Skp2 in lymphomagenesis. Proc. Natl Acad. Sci. USA 98, 2515–2520 (2001).
De Brakeleer, S. et al. Cancer predisposing missense and protein truncating BARD1 mutations in non-BRCA1 or BRCA2 breast cancer families. Hum. Mutat. 31, e1175–e1185 (2010).
Ratajska, M. et al. Cancer predisposing BARD1 mutations in breast-ovarian cancer families. Breast Cancer Res. Treat. 23 Feb 2011 (doi:10.1007/s10549-011-1403-8).
Sabatier, R. et al. BARD1 homozygous deletion, a possible alternative to BRCA1 mutation in basal breast cancer. Genes Chromosomes Cancer 49, 1143–1151 (2010).
Ruffner, H., Joazeiro, C. A., Hemmati, D., Hunter, T. & Verma, I. M. Cancer-predisposing mutations within the RING domain of BRCA1: loss of ubiquitin protein ligase activity and protection from radiation hypersensitivity. Proc. Natl Acad. Sci. USA 98, 5134–5139 (2001).
Dias, D. C., Dolios, G., Wang, R. & Pan, Z. Q. CUL7: a DOC domain-containing cullin selectively binds Skp1·Fbx29 to form an SCF-like complex. Proc. Natl Acad. Sci. USA 99, 16601–16606 (2002).
Skaar, J. R. et al. PARC and CUL7 form atypical cullin RING ligase complexes. Cancer Res. 67, 2006–2014 (2007).
Kim, S. S. et al. CUL7 is a novel antiapoptotic oncogene. Cancer Res. 67, 9616–9622 (2007).
Oliner, J. D., Kinzler, K. W., Meltzer, P. S., George, D. L. & Vogelstein, B. Amplification of a gene encoding a p53-associated protein in human sarcomas. Nature 358, 80–83 (1992).
Bo, M. D. et al. MDM4 (MDMX) is overexpressed in chronic lymphocytic leukaemia (CLL) and marks a subset of p53wild-type CLL with a poor cytotoxic response to Nutlin-3. Br. J. Haematol. 150, 237–239 (2010).
Danovi, D. et al. Amplification of Mdmx (or Mdm4) directly contributes to tumor formation by inhibiting p53 tumor suppressor activity. Mol. Cell. Biol. 24, 5835–5843 (2004).
Riemenschneider, M. J. et al. Amplification and overexpression of the MDM4 (MDMX) gene from 1q32 in a subset of malignant gliomas without TP53 mutation or MDM2 amplification. Cancer Res. 59, 6091–6096 (1999).
Nikolaev, A. Y., Li, M., Puskas, N., Qin, J. & Gu, W. Parc: a cytoplasmic anchor for p53. Cell 112, 29–40 (2003).
Duan, W. et al. Expression of Pirh2, a newly identified ubiquitin protein ligase, in lung cancer. J. Natl. Cancer Inst. 96, 1718–1721 (2004).
Leng, R. P. et al. Pirh2, a p53-induced ubiquitin-protein ligase, promotes p53 degradation. Cell 112, 779–791 (2003).
Hu, J. et al. WD40 protein FBW5 promotes ubiquitination of tumor suppressor TSC2 by DDB1–CUL4–ROC1 ligase. Genes Dev. 22, 866–871 (2008).
Figueroa, A. et al. Novel roles of hakai in cell proliferation and oncogenesis. Mol. Biol. Cell 20, 3533–3542 (2009).
Annunziata, C. M. et al. Frequent engagement of the classical and alternative NF-κB pathways by diverse genetic abnormalities in multiple myeloma. Cancer Cell 12, 115–130 (2007).
Danson, S., Dean, E., Dive, C. & Ranson, M. IAPs as a target for anticancer therapy. Curr. Cancer Drug Targets 7, 785–794 (2007).
Huang, H. et al. The inhibitor of apoptosis, cIAP2, functions as a ubiquitin-protein ligase and promotes in vitro monoubiquitination of caspases 3 and 7. J. Biol. Chem. 275, 26661–26664 (2000).
Keats, J. J. et al. Promiscuous mutations activate the noncanonical NF-κB pathway in multiple myeloma. Cancer Cell 12, 131–144 (2007).
Yang, Y., Fang, S., Jensen, J. P., Weissman, A. M. & Ashwell, J. D. Ubiquitin protein ligase activity of IAPs and their degradation in proteasomes in response to apoptotic stimuli. Science 288, 874–877 (2000).
Compagno, M. et al. Mutations of multiple genes cause deregulation of NF-κB in diffuse large B-cell lymphoma. Nature 459, 717–721 (2009).
Gemmill, R. M. et al. The hereditary renal cell carcinoma 3;8 translocation fuses FHIT to a patched-related gene, TRC8. Proc. Natl Acad. Sci. USA 95, 9572–9577 (1998).
Gimelli, S. et al. The tumor suppressor gene TRC8/RNF139 is disrupted by a constitutional balanced translocation t(8;22)(q24.13;q11.21) in a young girl with dysgerminoma. Mol. Cancer 8, 52 (2009).
Lee, J. P. et al. The TRC8 ubiquitin ligase is sterol regulated and interacts with lipid and protein biosynthetic pathways. Mol. Cancer Res. 8, 93–106 (2010).
Tsai, Y. C. et al. The ubiquitin ligase gp78 promotes sarcoma metastasis by targeting KAI1 for degradation. Nature Med. 13, 1504–1509 (2007).
Acknowledgements
The authors apologize to the many scientists whose contributions to this extensive and exponentially growing field of research could not be directly cited because of space limitations. They are indebted to their colleagues whose outstanding reviews on individual RING finger E3s and RING finger E3 families summarize and cite much of this important research. The authors thank R. Das (Structural Biophysics Laboratory, National Cancer Institute, USA) for invaluable assistance in generating the RING finger-E2 ribbon diagram. The authors' research programmes are supported by the National Institutes of Health, National Cancer Institute, Center for Cancer Research, USA.
Author information
Authors and Affiliations
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Related links
Supplementary information
Supplementary information S1 (table)
Established and Proposed E3s for p53 (PDF 385 kb)
Supplementary information S2 (table)
RING Finger E3s Implicated in Regulating Cell Cycle in Response to DNA Damage (PDF 173 kb)
Supplementary information S3 (table)
RING E3s Implicated in DNA repair* (PDF 187 kb)
Glossary
- Oncogenes
-
Genes with protein products that can promote cancer development. Oncogenes frequently undergo amplification or activating mutations and act in a genetically dominant manner.
- Tumour suppressor genes
-
(TSGs). Genes whose protein products, when lost or mutated, are permissive for the development of cancer. TSGs frequently undergo deletion or inactivating mutations of both alleles and act in a genetically recessive manner.
- Aneuploidy
-
Abnormal number of chromosomes resulting in more or less than the normal diploid number of chromosomes. Cancer cells are frequently aneuploid.
- Cyclins
-
Proteins that control the progression of the cell cycle by activating cyclin-dependent kinases.
- Securin
-
A protein that forms a complex with separase and thereby inhibits separase activity and prevents chromosome separation at anaphase. Securin is dephosphorylated and degraded by APC/CCDC20 at the onset of anaphase.
- Haploinsufficient TSG
-
A gene for which the loss of one allele is sufficient to promote cancer development.
- Amplification
-
Increased copy number of a gene within the genome; this is a common mechanism to increase the activity of oncogenes.
- Anergy
-
When lymphocytes fail to mount an immune response to an antigen.
Rights and permissions
About this article
Cite this article
Lipkowitz, S., Weissman, A. RINGs of good and evil: RING finger ubiquitin ligases at the crossroads of tumour suppression and oncogenesis. Nat Rev Cancer 11, 629–643 (2011). https://doi.org/10.1038/nrc3120
Published:
Issue Date:
DOI: https://doi.org/10.1038/nrc3120
This article is cited by
-
UBE4B interacts with the ITCH E3 ubiquitin ligase to induce Ku70 and c-FLIPL polyubiquitination and enhanced neuroblastoma apoptosis
Cell Death & Disease (2023)
-
Oral squamous cell carcinomas: state of the field and emerging directions
International Journal of Oral Science (2023)
-
A prospective prognostic signature for pancreatic adenocarcinoma based on ubiquitination-related mRNA-lncRNA with experimental validation in vitro and vivo
Functional & Integrative Genomics (2023)
-
RIG-I promotes cell proliferation in esophageal squamous cell carcinoma by facilitating p21 degradation
Medical Oncology (2023)
-
Identifying common transcriptome signatures of cancer by interpreting deep learning models
Genome Biology (2022)