Abstract
Statins reduce cardiovascular morbidity and mortality in appropriately selected patients. However, statin-associated myopathy is a significant risk associated with these agents. Recently, variation in the SLCO1B1 gene was reported to predict simvastatin-associated myopathy. The aim of this study was to replicate association of the rs4149056 variant in SLCO1B1 with severe statin-associated myopathy in a cohort of patients using a variety of statin medications and to investigate the association with specific statin types. We identified 25 cases of severe statin-associated myopathy and 84 controls matched for age, gender, statin type and dose. The rs4149056 variant in SLCO1B1 was not significantly associated with myopathy in this group as a whole. However, when subjects were stratified by statin type, the SLCO1B1 rs4149056 genotype was significantly associated with myopathy in patients who received simvastatin, but not in patients who received atorvastatin. Our findings provide further support for a role for SLCO1B1 genotype in simvastatin-associated myopathy, and suggest that this association may be stronger for simvastatin compared with atorvastatin.
Similar content being viewed by others
Introduction
Despite their significant effects on reducing cardiovascular mortality, statins remain underused in at-risk patients,1 in part because of concerns of adverse drug reactions affecting the musculoskeletal system. Statins are associated with a spectrum of muscle pain and damage varying in severity and frequency. At one end of this spectrum is rhabdomyolysis, a rare but potentially fatal complication that occurs in fewer than one in 10 000 patients receiving statins per year.2 The devastating consequences of statin-induced rhabdomyolysis led to the withdrawal of cerivastatin in 2001 after ∼100 rhabdomyolysis-related deaths were associated with this drug.3 Statin-associated myopathy, defined as elevation of creatine kinase (CK) values greater than 10 times the upper limit of normal and often accompanied by muscle pain, occurs in up to 0.4% of patients on high doses of simvastatin4 and in 0.01–0.1% of patients on more standard doses.5, 6 Less severe muscle pain without biochemical evidence of muscle damage is a common complaint and occurs in up to 5–10% of patients on statins in observational studies.7, 8 In a systematic review of randomized trials comparing statin therapy with placebo, the risk of myalgia was not significantly increased in patients administered statins.9
The mechanisms underlying statin-associated myopathy remain incompletely understood and methods to identify patients at high risk are lacking. Recently, genetic variants in the gene encoding the organic anion-transporting poly-peptide OATP1B1 (gene name SLCO1B1, GeneID: 10599) were reported to be significantly associated with myopathy in two cohorts of patients from the United Kingdom receiving high doses of simvastatin.10 In patients with ‘definite myopathy’, defined as a CK value greater than 10 times the upper limit of normal, the minor allele of the rs4149056 single nucleotide polymorphism (SNP) in SLCO1B1 (SLCO1B1*5 genotype), which results in the substitution of alanine for valine at amino acid residue 174, conferred an allelic odds ratio (OR) of 4.5. This variant has also been reported to be associated with more mild simvastatin-associated adverse effects consisting of muscle pain, drug discontinuation or CK elevation greater than three times the upper limit of normal with an OR of 1.7 per SLCO1B1 genotype.11 However, no replication has been reported for the association of this variant with significant, biochemically determined myopathy and, in addition, the role of this variant in myopathy associated with other statins has not been explored. Here we investigate the role of the rs4149056 SNP in SLCO1B1 in a cohort of European patients with statin-associated myopathy. This study was designed to replicate the previously reported association of rs4149056 with severe myopathy and to investigate the statin-specificity of this association.
Patients and methods
Patients
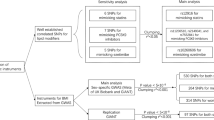
Cases were identified by searching the medical records of two large lipid clinics in the Netherlands (Amsterdam and Slotervaartziekenhuis). As muscle symptoms are an insensitive marker of myopathy and rhabdomyolysis12 and were inconsistently reported in the clinical records, we used a biochemical definition of myopathy consisting of plasma creatine kinase values greater than ten times the upper limit of normal for the reference laboratory (150 U l−1), as was used in the SEARCH study.10 Patients with CK elevations that occurred in the setting of an acute coronary syndrome or myocardial infarction were excluded. For each case, we identified between two and four control patients matched for age, gender, statin type and dose. All patients were of Dutch ancestry. This study was approved by the ethical review boards of the University of Amsterdam and the University of British Columbia.
Genotyping
DNA extraction and genotyping were performed as previously described.13 We designed an Illumina Goldengate genotyping assay using a custom oligo pool for the rs4149056 variant at the SLCO1B1 locus. All SNP genotype data were manually clustered using BeadStudio software (Illumina, San Diego, CA, USA). The average genotyping call rate for all samples was >98%.
Power calculation and statistical analysis
We calculated that to identify an association of the rs4149056 SNP with myopathy with an effect size of 3.0 or greater (similar to that observed in the SEARCH study) would require at least 20 cases assuming a similar minor allele frequency. The primary end point of this study was replication of the association of rs4149056 with myopathy in an appropriately powered sample of patients using a variety of statins. Secondary end points were the association of myopathy with rs4149056 in patients taking specific statin types. Genotype frequencies in cases and controls were compared using a genotypic dominant model. As this was designed as a replication study under the previous hypothesis that the rs4149056 minor allele frequency would be higher in cases than controls, we used a 1-tailed P value. Under the assumptions of this test a finding of the rs4149056 risk allele being more frequent in controls than cases would be considered erroneous. A P value <0.05 was considered significant.
Results
We examined the records of nearly 9000 patients from two lipid clinics in the Netherlands. Twenty-five patients with statin-associated myopathy, defined as a CK>1500 U l−1 were identified, yielding a prevalence of 0.26%, similar to rates reported for equivalently defined phenotypes.5, 10, 14 For each case we identified 2–4 controls matched for age, gender, statin type and dose. The clinical characteristics of the cases and controls are shown in Table 1. There were no significant differences between cases and controls in age or gender. Most patients received simvastatin or atorvastatin, and the distribution of patients on each statin and total dose did not differ between groups. More cases than controls received a concomitant fibrate medication, whereas the use of calcium channel antagonists and niacin was more common in controls. These differences did not reach statistical significance. No cases or controls received amiodarone. Overall, 32.0% of cases and 28.6% of controls received one of these four additional medications (χ2 P value=0.74). No cases were reported to have received other known inhibitors of CYP3A4 or SLCO1B1 (protease inhibitors, macrolide antibiotics, azole antifungals, thiazolidinediones, tacrolimus, cylcosporin or rifampicin).
Amongst the entire study group, there was a non-significant trend towards increased risk of myopathy with SLCO1B1 risk genotype (TC or CC vs TT genotype OR 1.5, 95% confidence interval (CI) 0.58–3.69, χ2 P=0.21, Fisher's exact P=0.14). The frequency of the rs4149056 risk ‘C’ allele was similar in cases (23%) and controls (19%). Therefore, in this cohort of patients on a variety of statins, the rs4149056 variant was not significantly associated with myopathy.
We next examined only those patients who received simvastatin, consisting of 12 cases and 39 controls. In this subset there were no significant differences in age, gender or simvastatin dose. In this group, SLCO1B1 rs4149056 genotype conferred a significant threefold increased risk for myopathy (OR 3.2, 95% CI 0.83–11.96, χ2 P=0.042, Fisher's exact P=0.064). The frequency of the ‘C’ allele was 33% in cases and 18% in controls. The association of rs4149056 risk genotype with myopathy remained significant after excluding those patients who received a fibrate, calcium antagonist or niacin (OR 4.5, 95% CI 0.73–27.59, χ2 P=0.044, Fisher's exact P=0.085) (Table 2).
In contrast to the results for simvastatin, in the limited number of patients receiving atorvastatin (10 cases and 35 controls) the SLC01B1 genotype conferred no increased risk of myopathy (OR 1.06, 95% CI 0.22–4.80, χ2 P=0.48, Fisher's exact P=0.30). The frequency of the ‘C’ allele did not differ between cases and controls (20 and 19%, respectively). These data suggest that the increased risk of myopathy associated with the rs4149056 genotype may be stronger for simvastatin than for other statins in this cohort (Figure 1).
SLCO1B1 rs4149056 genotype influences susceptibility to myopathy in response to simvastatin but not atorvastatin. ORs and 95% CIs for myopathy with SLCO1B1 rs4149056 risk genotype are shown per statin type. In the entire cohort of patients a nonsignificant trend to increased myopathy risk was observed in individuals with the risk ‘C’ allele. In the subset of patients administered simvastatin, rs4149056 genotype conferred a threefold increased risk for myopathy (One-tailed χ2 P value 0.042). In contrast, in patients administered atorvastatin rs4149056 conferred no increased risk for myopathy, suggesting that the influence of SLCO1B1 on myopathy is statin specific.
Discussion
Statin-associated myopathy is a rare but clinically important adverse drug reaction. Currently we lack the ability to identify patients at high risk of developing this complication. Recently, variants in the SLCO1B1 gene were identified as a significant risk factor for simvastatin-associated myopathy.10 We identified patients from two large lipid clinics with an extreme phenotype consisting of CK elevations 10 times greater than the upper limit of normal. We chose a biochemical rather than a clinical phenotype because the association between muscle symptoms and objective findings of muscle damage is weak. Our results confirm that the rs4149056 variant in SLCO1B1 influences simvastatin-associated myopathy, and suggest that this association is less significant for atorvastatin.
Based on our power calculations, we had greater than 80% power to identify an association of rs4149056 with myopathy in the entire cohort with an effect size of 3.0 or greater. Therefore, the lack of association of this SNP with myopathy in a cohort of patients on a variety of statins is unlikely to represent type II error. This finding indicates that rs4149056 is unlikely to influence myopathy in response to statins in general. In contrast, the sample of 10 cases and 34 controls using atorvastatin provides only 44–52% power to detect an association with myopathy with an effect size of 3–3.5, as we observed for simvastatin. In addition, the CIs for patients administered simvastatin and atorvastatin were overlapping. Therefore, although our results do not support an association between rs4149056 and atorvastatin-associated myopathy, our ability to refute such an association is limited due to the relatively small number of patients with this extreme phenotype, and this finding should be considered hypothesis generating. In patients who received simvastatin, the OR of myopathy of 3.2 per genotype, or 2.3 per ‘C’ allele, is lower than that reported in the SEARCH study (4.5 per ‘C’ allele) but similar to that in the replication Heart Protection Study Cohort (2.6 per ‘C’ allele),10 suggesting that the true magnitude of the effect of SLCO1B1 is likely within this range. The mean simvastatin dose in our population (∼30 mg) was lower than that in the SEARCH study (80 mg) and Heart Protection study (40 mg) and our findings therefore extend this association to lower doses of this medication.
SLCO1B1 mediates the transport of statins into hepatocytes, including simvastatin and atorvastatin.15 The frequency of the rs4149056 minor allele ranges from <1% in Yoruba Nigerians to more than 20% in European populations (Hapmap data). The valine to alanine substitution at amino acid 174 that is introduced by this SNP appears to be a functionally important variant, as individuals heterozygous or homozygous for this SNP have increased concentrations of drug in plasma after oral administration of a variety of statins.16, 17 In addition, in transfected HeLa cells, the rs4149056 polymorphism results in reduced substrate transport,18 further supporting the concept that this is a low activity allele resulting in diminished statin uptake by hepatocytes. Therefore, it seems likely that this SNP directly impacts simvastatin-associated myopathy, potentially via an effect on plasma drug concentrations, and is not merely a marker for a nearby variant in linkage disequilibrium with rs4149056.
The impact of the rs4149056 variant on statin metabolism appears to differ between different statins. Pasanen et al. administered oral simvastatin acid, atorvastatin and rosuvastatin to healthy volunteers and compared the area under the plasma concentration time curve (AUC) between individuals based on their rs4149056 genotypes.16, 17 For simvastatin acid, the AUC was increased by 221% in genotype CC individuals (n=4) compared with wild-type TT individuals (n=16). For atorvastatin, AUC was increased 145% and for rosuvastatin 62% (One-way analysis of variance P<0.05 for difference in AUC between three statins). These data indicate that the effect of rs4149056 is greatest for simvastatin and less for atorvastatin and rosuvastatin, and provide an in vivo pharmacokinetic correlate for the differential pharmacogenetic risk we observed between simvastatin and atorvastatin. Rosuvastatin has been associated with a higher risk of myopathy compared with other statins in some studies.19 Only one patient in our cohort received rosuvastatin, but the available pharmacokinetic data predict that myopathy associated with this drug would be less influenced by rs4149056 genotype. A stronger effect of rs4149056 genotype in patients with simvastatin compared with atorvastatin has also been reported for patients with milder symptoms consisting of subjective reports of myalgias, drug discontinuation, or CK values greater than three times the upper limit of normal.11
In summary, our results provide further support for the important role of SLCO1B1 in influencing simvastatin-related myopathy. Importantly, our data suggest that this association may not exist or may be weaker for other statins, and therefore raise the possibility that patients found to be at high risk for simvastatin-associated myopathy by virtue of their SLCO1B1 genotype may be at lower risk for myopathy if treated with an alternative statin.
References
Mitka M . Expanding statin use to help more at-risk patients is causing financial heartburn. J Am Med Assoc 2003; 290: 2243–2245.
Silva MA, Swanson AC, Gandhi PJ, Tataronis GR . Statin-related adverse events: a meta-analysis. Clin Ther 2006; 28: 26–35.
Staffa JA, Chang J, Green L . Cerivastatin and reports of fatal rhabdomyolysis. N Engl J Med 2002; 346: 539–553a.
de Lemos JA, Blazing MA, Wiviott SD, Lewis EF, Fox KAA, White HD et al. Early intensive vs a delayed conservative simvastatin strategy in patients with acute coronary syndromes: phase Z of the A to Z trial. J Am Med Assoc 2004; 292: 1307–1316.
Dujovne CA, Chremos AN, Pool JL, Schnaper H, Bradford RH, Shear CL et al. Expanded clinical evaluation of lovastatin (EXCEL) study results: IV. Additional perspectives on the tolerability of lovastatin. Am J Med 1991; 91 (1B): 25S–30S.
Law M, Rudnicka AR . Statin safety: a systematic review. Am J Cardiol 2006; 97 (Suppl 1): S52–S60.
Bruckert E, Hayem G, Dejager S, Yau C, Begaud B . Mild to moderate muscular symptoms with high-dosage statin therapy in hyperlipidemic patients--the PRIMO study. Cardiovasc Drugs Ther 2005; 19: 403–414.
Nichols GA, Koro CE . Does statin therapy initiation increase the risk for myopathy? An observational study of 32 225 diabetic and nondiabetic patients. Clin Ther 2007; 29: 1761–1770.
Kashani A, Phillips CO, Foody JM, Wang Y, Mangalmurti S, Ko DT et al. Risks associated with statin therapy: a systematic overview of randomized clinical trials. Circulation 2006; 114: 2788–2797.
The SEARCH Collaborative Group. SLCO1B1 variants and statin-induced myopathy—a genomewide study. N Engl J Med 2008; 359: 789–799.
Voora D, Shah SH, Spasojevic I, Ali S, Reed CR, Salisbury BA et al. The SLCO1B1*5 genetic variant is associated with statin-induced side effects. J Am Coll Cardiol 2009; 54: 1609–1616.
Mohaupt MG, Karas RH, Babiychuk EB, Sanchez-Freire V, Monastyrskaya K, Iyer L et al. Association between statin-associated myopathy and skeletal muscle damage. CMAJ 2009; 181: E11–E18.
Ross CJD, Katzov-Eckert H, Dube MP, Brooks B, Rassekh SR, Barhdadi A et al. Genetic variants in TPMT and COMT are associated with hearing loss in children receiving cisplatin chemotherapy. Nat Genet 2009; 41: 1345–1349.
Tobert JA . Efficacy and long-term adverse effect pattern of lovastatin. Am J Cardiol 1988; 62: 28J–34J.
Ghatak A, Faheem O, Thompson PD . The genetics of statin-induced myopathy. Atheroscler 2009; 210: 337–343.
Pasanen MK, Fredrikson H, Neuvonen PJ, Niemi M . Different effects of SLCO1B1 polymorphism on the pharmacokinetics of atorvastatin and rosuvastatin. Clin Pharmacol Ther 2007; 82: 726–733.
Pasanen MK, Neuvonen M, Neuvonen PJ, Niemi M . SLCO1B1 polymorphism markedly affects the pharmacokinetics of simvastatin acid. Pharmacogenet Genomics 2006; 16: 873–879.
Tirona RG, Leake BF, Merino G, Kim RB . Polymorphisms in OATP-C. J Biol Chem 2001; 276: 35669–35675.
Davidson MH, Clark JA, Glass LM, Kanumalla A . Statin safety: an appraisal from the adverse event reporting system. Am J Cardiol 2006; 97 (Suppl 1): S32–S43.
Acknowledgements
This work was supported by grants from the Canadian Institutes of Health Research and the Canadian Foundation for Innovation. LRB is a Canadian Pharmacogenomic Network for Drug Safety fellow and received support from the UBC Department of Medicine. HV is supported by fellowships from the Michael Smith Foundation for Health Research and Child and Family Research Institute.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
PowerPoint slides
Rights and permissions
This work is licensed under the Creative Commons Attribution-NonCommercial-No Derivative Works 3.0 Unported License. To view a copy of this license, visit http://creativecommons.org/licenses/by-nc-nd/3.0/
About this article
Cite this article
Brunham, L., Lansberg, P., Zhang, L. et al. Differential effect of the rs4149056 variant in SLCO1B1 on myopathy associated with simvastatin and atorvastatin. Pharmacogenomics J 12, 233–237 (2012). https://doi.org/10.1038/tpj.2010.92
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/tpj.2010.92
Keywords
This article is cited by
-
Frequency of functional exonic single-nucleotide polymorphisms and haplotype distribution in the SLCO1B1 gene across genetic ancestry groups in the Qatari population
Scientific Reports (2022)
-
A systematic review and meta-analysis of genotype-based and individualized data analysis of SLCO1B1 gene and statin-induced myopathy
The Pharmacogenomics Journal (2021)
-
Correlation between single-nucleotide polymorphisms and statin-induced myopathy: a mixed-effects model meta-analysis
European Journal of Clinical Pharmacology (2021)
-
The atorvastatin metabolic phenotype shift is influenced by interaction of drug-transporter polymorphisms in Mexican population: results of a randomized trial
Scientific Reports (2020)
-
The impact of statins on physical activity and exercise capacity: an overview of the evidence, mechanisms, and recommendations
European Journal of Applied Physiology (2020)