Abstract
More than 95% of genes in the human genome are alternatively spliced to form multiple transcripts, often encoding proteins with differing or opposing function. The control of alternative splicing is now being elucidated, and with this comes the opportunity to develop modulators of alternative splicing that can control cellular function. A number of approaches have been taken to develop compounds that can experimentally, and sometimes clinically, affect splicing control, resulting in potential novel therapeutics. Here we develop the concepts that targeting alternative splicing can result in relatively specific pathway inhibitors/activators that result in dampening down of physiologic or pathologic processes, from changes in muscle physiology to altering angiogenesis or pain. The targets and pharmacology of some of the current inhibitors/activators of alternative splicing are demonstrated and future directions discussed.
I. Introduction
The human genome generates up to a million different proteins from a genetic code that contains just over 20,000 distinct genes. There is therefore a substantial increase in diversity between the genetic code and the proteome that functions to control how cells behave, interact, and form complex tissues and organs. One of the principal mechanisms of this diversity is the generation of multiple mRNA splice forms from a single genetic unit by the process of alternative splicing. Although this process has been known about for nearly 50 years, the mechanisms underlying its control in health and disease are only now becoming understood sufficiently well that intervention in the process can be considered as a potential therapeutic approach. Over the last few years a number of pharmacological strategies have been developed to target alternative splicing in disease states, and some of these have now reached the clinical trial stage, with surprising, sometimes promisingly efficacious, results. However, the generation of new drugs that target splicing raises a number of challenges that are relatively restricted to this type of agent. These challenges, both real and perceived, need to be addressed when developing such agents. The potential for novel, potent, efficacious therapeutics in this field is clearly enormous, but the barriers to successful drug development need to be understood, negotiated, overcome, and sometimes eliminated for this potential to be realized. A comprehension of the principles of the alternative splicing process, its regulation, and the concepts that underlie coordinated alternative splicing is required so that common initial concerns about these pharmacological approaches can be relieved. In this review we intend to layout the principles behind the development of approaches to regulate alternative splicing in health and disease.
II. Methods
Searches of PubMed, Web of Science, and clinicaltrials. gov were used to identify papers, citations, and clinical trials, respectively, which addressed the field of splicing and development of novel therapeutics. Background information was gleaned from recent reviews, textbooks, and integrated from the authors’ own knowledge of the systems. Search terms in PubMed used to identify primary sources included: Alternative Splicing AND therapeutics (716 hits) and Alternative Splicing AND clinical trial (86 hits). From these searches, areas of disease and mechanisms of action were further explored, including searches on cancer, apoptosis, angiogenesis, pain, muscular dystrophy, and then drilled down in terms of mechanisms and target molecules. This was followed up by searches using clinicaltrials.gov, Google, and Web of Science to identify additional information about the targets.
III. Alternative Splicing
Alternative splicing is the process of combinatorial rearrangement of exons, parts of exons, and/or parts of introns into mature RNA to result in a multitude of transcripts. It has only recently been shown to be extensive across the human genome, with more than 94% of genes being alternatively spliced (Pan et al., 2008; Wang et al., 2008). This makes alternative splicing one of the main drivers of proteome diversity and therefore a major regulatory level for cell functions. Interestingly, although conservation at the DNA sequence level is quite high in vertebrates, alternative splicing (AS) is less conserved in the evolutionary tree, with a degree of similarity between human and mouse of only ∼30% (Barbosa-Morais et al., 2012).
The spliceosome, a macromolecular complex formed of many proteins as well as small nuclear ribonucleoproteins, is the machinery that performs the removal of regions of the RNA – introns - and joining of regions destined to form mature mRNA – exons (Will and Lührmann, 2011). It does this by assembling at splice sites, regions at the junction between exons and introns, which are fairly well conserved. Through complex binding of the proteins and RNA in the spliceosome with the target RNA sequence, the introns are spliced out with the occurrence of two trans-esterification reactions. This involves the upstream and downstream intronic splice sites – so-called 5′ and 3′ splice sites – and a conserved sequence on the intron called the branch point. A region rich in pyrimidine nucleotides (C and T) helps the spliceosome assemble through binding of various splice factors and is also one of the main features of a basic splicing reaction (Fig. 1).
Control of splicing. Cis acting elements, such as exon splicing enhancer sequences (ESE), exon splicing silencers (ESS), intron splicing silencers (ISS), and intron splicing enhancers (ISE) are bound by trans-acting elements (splicing factors, SF) that facilitate or repress splicing. At the 3′ end of the intron, a consensus AG splice site is preceded by a polypyrimidine (pY) tract and a branch point that requires a consensus splice site (YNYURAY, R = purine, U = uridine). The complex of factors bound to the 5′ splice site (gray region) interacts with the complex bound to the 3′ site (gray region) to induce and regulate splicing.
There are several modes of alternative splicing: an exon may be included or excluded in the final transcript—known as a cassette exon; two exons may be alternatively included or skipped—mutually exclusive exons; alternatively, 5′ or 3′ splice sites may be used or a whole intron may be retained. These rearrangements may occur either in the coding or noncoding region of the mRNA. When occurring in the 5′ or 3′ untranslated regions it does not affect the protein sequence but may regulate its expression.
Analogous to transcription, alternative splicing is regulated by both cis- and trans-acting elements. Both introns and exons have regions that may affect the inclusion/exclusion of certain exons—cis-acting elements found on the RNA. Depending on whether they are inhibitory or stimulatory for certain splicing events, they are termed exonic splicing silencer or enhancer (ESS or ESE) and intronic splicing silencer or enhancer (ISS or ISE) sequences (Fig. 1). They often work through binding of trans-acting elements, often proteins called splice factors (SF). Several RNA-binding proteins are classified as SFs. Two of the major classes are SR proteins (containing serine arginine-rich motifs) and heterogeneous nuclear ribonucleoproteins (hnRNPs). Classically, SR proteins were thought as activators and hnRNPs as silencers of AS; however, recent evidence shows that depending on the context each can act both ways (Fu and Ares, 2014). An increasing number of RNA binding proteins are also characterized as SFs outside of these major classes, many of them regulating cell- and tissue-specific splicing events [e.g., epithelial splicing regulatory proteins (ESRP) 1 and 2, CELF (cytidine-uridine-guanidine binding protein and Elav-like family), RbFox1 and 2, Muscleblind like (Mbnl)] (Kalsotra et al., 2008; Warzecha et al., 2009; Singh et al., 2014; Pedrotti et al., 2015).
IV. Targets within Alternative Splicing
The regulation of alternative splicing can be targeted therapeutically by devising agents that interfere either with the RNA
As it is transcribed and screened by the splicing machinery;
By modifying and preventing RNA binding proteins from recognizing their RNA targets or their actions on the splicing machinery; or
By regulating the controlling agents that allow the RNA binding proteins to carry out their splicing actions.
A. Nucleotide Binding Agents
To complete splicing at a specific point in the RNA sequence, the splicing factors need to be able to recognize specific sequences. There have been a number of attempts to define consensus sequences for various splicing proteins, and the most widely studied now have semispecific sequences that can be searched for to investigate splice sites. However, RNA splicing sequences are not as fixed as, for instance, DNA transcription factor consensus sequences (De Conti et al., 2013), and the ability of splicing factors to bind to their consensus sequences is clearly highly dependent on local secondary and tertiary RNA structure (Rosonina and Blencowe, 2004), binding with other proteins (Bradley et al., 2015), and many other factors and processes that take place within the cells. Despite this, it has been possible to generate short sequences of RNA that can specifically interfere with alternative splicing. There have been two main approaches:
Design complementary nucleotide sequences to specific splice regions such that splicing is skipped. These dominant negative splicing inhibitors have taken the form of either RNA or modified RNAs that can interfere with specific targeted sequences. For example, in spinal muscular atrophy (SMA) (Osman et al., 2014; Rigo et al., 2014) apoptosis, DNA repair response (MDM4 function) (Dewaele et al., 2016; Li et al., 2016), and in other conditions.
RNA sequences can be used that are targets for specific splicing factors that can then be used to position the splicing factors at a specific splice point or mop up splicing factors so that they cannot act on their targets. One example is targeted oligonucleotide enhancers of splicing (Smith et al., 2014) and another is to use alternative intronic splicing silencers [ISS (Singh et al., 2015)], which can then be used to stimulate or repress splicing to a specific exon, respectively.
B. RNA Binding Agents
A growing number of RNA binding proteins, such as the classic SR proteins and the proteins that form the basis of the splicing factor machinery, have been identified as taking part in splicing—alternative or constitutive—and it has been found that modulation of these can result in changes in expression of splice isoforms. For example, small molecule inhibitors of SRSF1 have been developed (Bakkour et al., 2007), whereas the SF3B family of proteins have been targeted by a number of compounds such as spliceostatin (Kaida et al., 2007) and pladienolide B (Kotake et al., 2007). A recent approach has been to use a generic screen, whereby alternative splicing constructs are used to screen chemical libraries that can identify compounds that alter alternative splicing in a nonbiased manner—i.e., not depending on the mechanism of action (Naryshkin et al., 2014; Palacino et al., 2015). This has resulted in a number of compounds where the mechanisms of action are unknown or only postulated [e.g., sudemycins, postulated to be through SF3b (Fan et al., 2011), or aryl-thiazol-piperidines, where no mechanism of action is known (Fan et al., 2011) and may not even be splicing modulators but differential posttranscriptional regulators].
C. Splicing Factor Regulatory Proteins
Splicing occurs, and alternative splicing is regulated, by the interaction of a host of proteins that form the spliceosome but do not directly bind to RNA and by proteins that regulate RNA binding proteins. These include the structural proteins at the heart of the spliceosome, such as SF3B complex or formin binding protein 21 (FBP21) (Bedford et al., 1998). It is possible to switch splicing by designing synthetic, or discovering naturally occurring, compounds that can interfere with this process. One example is the discovery of borrelidin, an antifungal compound, modifications of which have been found to have antiangiogenic activity in tumors and which bind to the splicing protein FBP21(Woolard et al., 2011). The borrelidin analog BC194 appears to be able to coordinate splicing of the vascular endothelial growth factor (VEGF) gene to skip the normal 3′ splice site used in tumor cells to use an alternate downstream 3′ splice site, resulting in a protein with an alternate C terminus and that prevents angiogenesis in vitro and in vivo. As the mechanism through which FBP21 binds to the splice factors that control VEGF are unknown, it may also affect splicing of a number of other genes that control angiogenesis, but its ability to bind FBP21 places it firmly in the category of splicing inhibitors (Woolard et al., 2011).
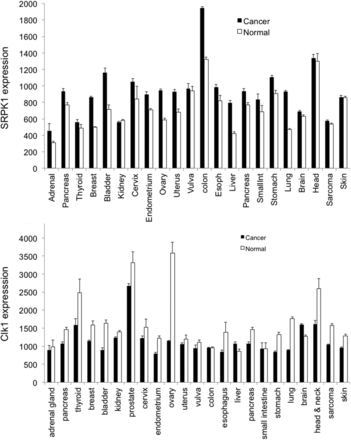
A second example is the use of compounds that inhibit splicing factor regulators, such as the splicing factor kinase family. These kinases are a small family of proteins that phosphorylate splicing factors of the SRSF1 family and include SRPK1, SRPK2, CLK1-4, DYRK1-2, PIM1-2, and PRP4. Inhibition of phosphorylation of the splicing factor SRSF1, for example, resulting in altered splicing of the downstream targets MKNK2, hnRNPA2/B1, and VEGF, all of which result in splice forms that switch from protumorigenic [MKNK2b contributes to the ras-mitogen-activated to mitogen activated protein kinases (MAPK) pathway by phosphorylation of elongation initiation factor 4E, which is oncogenic (Maimon et al., 2014)]. VEGF-A165a is proangiogenic, and hnRNPA2 can regulate VEGF-A translation to prevent translational read-through and prevent expression of VEGFAx, an antiangiogenic isoform (Eswarappa et al., 2014). SRSF1 has been shown to be a potent protumor splicing factor (Karni et al., 2007). Studies overexpressing SRSF1 have identified over 300 alternative splice variants that can be regulated, many of which are thought to contribute to tumor growth (see below) (Anczukow et al., 2015). Interestingly, SRPK1 is overexpressed in many tumors (Fig. 2), in contrast to other splice factor kinases such as CLK1, which are downregulated. Moreover, SRPK1 has been shown to be a key regulator of melanoma (Gammons et al., 2014), acute myeloid leukemia (Siqueira et al., 2015); glioma (Wu et al., 2013); hepatocellular carcinoma (Zhou et al., 2013); and lung (Gout et al., 2012), breast, prostate (Mavrou et al., 2015), pancreatic (Hayes et al., 2006), colorectal (Hayes et al., 2007; Amin et al., 2011); and ovarian cancer (Odunsi et al., 2012). In fact SRPK1 expression is a predictive biomarker for all-cause mortality in breast cancer (Li et al., 2014) and, given the effect of upregulation of SRSF1 in this cancer type, it is perhaps not surprising that SRPK1 targeting is a potential novel therapeutic for cancer treatments.
Expression data from the GENT database of SRPK1 (A) and CLK1 (B) in different cancer types (relative to a normalized target density of 500, using Affymetric microarrays) (Shin et al., 2011).
V. Pathways Regulated by Alternative Splicing
The number of physiologic and pathologic processes that appear to be regulated by alternative splicing is increasing rapidly, and it is likely that alternative splicing can contribute to all of the different cell processes. However, some processes have been at the forefront of research into modulation of alternative splicing, and it is these where the most advanced therapeutic approaches have been developed. Although we focus here on apoptosis, angiogenesis, cancer, pain, and neuromuscular degeneration, it is likely that modifications in splicing in cognitive dysfunction (Scheckel et al., 2016), liver (Sehgal et al., 2013) and lung disease (Nance et al., 2014), inflammation (Hirata et al., 2015), and reproduction (Ahmad and Ahmed, 2004; Bills et al., 2009), among many other processes, will come to the fore over the next few years. However, we have focused on the more advanced examples to provide proof of principle for how the new pharmacology of splicing is being developed.
A. Apoptosis
Apoptosis, or programmed cell death, is an essential mechanism in cell physiology, which often is deregulated in disease. The molecules involved in both intrinsic and extrinsic apoptotic pathways have a large number of splice variants reported. Interestingly, in many cases these splice variants have antagonistic properties—i.e., pro- and antiapoptotic, which suggest that modulation of AS may be a quick and economic way for the cell to switch its apoptotic potential depending on the intra- and extracellular signals. A few examples are presented here.
Both caspase 8 and 9 have functionally different isoforms. Whereas caspase 8 is normally proapoptotic, an intron retention forms Cas8L with antiapoptotic properties (Himeji et al., 2002). Skipping of exons 3, 4, 5, and 6 in caspase 9 results in a shorter protein that also has antiapoptotic properties—this event being regulated by SRSF1 and SRSF2 (Shultz et al., 2010; Pan et al., 2011). Yet another type of AS—5′ alternative splice sites—is responsible for a proapoptotic isoform of the well-known survival factor Bcl-xL, termed Bcl-xS (Cloutier et al., 2008). The balance between these two isoforms is known to be regulated by SFs hnRNP A1 and H/F as well as signaling through PKC (Shultz et al., 2012). Alternate inclusion of exon 6 in the Fas receptor results in either the trans-membrane protein, which is proapoptotic, or a soluble receptor form with opposing functions (Bonnal et al., 2008).
A theme that is emerging in several other physiologic and pathologic processes, occurs in apoptosis too, coordinated regulation of several splice isoforms to define a specific cell function. Such is the example of the E2F1 transcription factor and the splice factor SRSF2 that coregulate splice isoforms of several apoptosis genes, c-flip, caspase 8, caspase 9, and Bcl-x (Merdzhanova et al., 2008).
B. Angiogenesis
VEGFs, particularly the isoform VEGF-A165a, are the principal angiogenic molecules in cancers and metastatic lesions(Hurwitz et al., 2004), blood vessel growth into the retina in blindness, and development of neovasculature in neointimal hyperplasia in heart disease, revascularization in peripheral and coronary vascular disease, and progression of arthritis, psoriasis, and many other diseases. The bioactivity of the VEGF ligands and receptors is regulated by alternative RNA splicing (Houck et al., 1992). VEGF-A is generated from a single gene by alternative splicing of eight exons (Harper and Bates, 2008). Exons 5, 6, and 7 are alternatively spliced to form isoforms of different lengths, with different heparin binding properties, with the 189 amino acid isoform being most heparin binding and the 110 or 121 amino acid isoforms having least heparin binding (Ferrara et al., 1991). Half of the isoforms, termed VEGF-Axxxa, where xxx refers to the number of amino acids, are generated using a proximal splice site in exon 8, resulting in a short open reading frame (6 amino acids) and a stop codon. The other half, the VEGF-Axxxb isoforms, use a distal splice site in exon 8, resulting in the same size open reading frame, and an alternate C terminus, which confers antiangiogenic properties (Bates et al., 2002). The most common of this family of splice variants (VEGF-A165b) is endogenously expressed in normal tissues and downregulated in colon (Varey et al., 2008), renal, and prostate cancer (Bates et al., 2002) and metastatic melanoma (Pritchard-Jones et al., 2007). Interestingly, anti-VEGF treatment, such as use of the antibody bevacizumab, acts on both isoforms. In a post hoc study in one of the registration trials of bevacizumab (ECOG E3200), relative VEGF-A165b expression predicted efficacy of bevacizumab treatment (Bates et al., 2012). There have therefore been clear demonstrations that VEGF splicing is critical not only for cancer growth but also to resistance to anti-VEGF treatment.
Mechanisms controlling splicing of exon 8 are beginning to be elucidated in model systems. SR proteins such as SRSF1, SRSF2, and SRSF6 regulate VEGF-A splicing; SRSF6 overexpression switches expression to VEGF-A165b and SRSF1 to VEGF-A165a in epithelial cells (Nowak et al., 2008) and in prostate (Mavrou et al., 2015), melanoma (Gammons et al., 2014), and colorectal (Amin et al., 2011) cancer cell lines. SRSF2 also regulates VEGF-A165b splicing in lung cancer cells (Merdzhanova et al., 2010). SRSF1 binding requires the 35 base region in intron 7 immediately upstream of the proximal splice site (VEGF-A165) (Nowak et al., 2010), whereas SRSF6 binding requires the 35 base region surrounding the distal splice site (Nowak et al., 2008). Inhibition of SRSF1 nuclear localization by blocking the SR protein kinase SRPK1 inhibits VEGF-A proximal splice site choice in exon 8 (Nowak et al., 2010) in all five cancer cell lines (two uveal and one cutaneous melanoma, PC3 prostate, and LS174t colorectal) and two epithelial cell lines (podocyte and retinal pigmented epithelial cells) so far investigated. However, regulation of VEGF splicing in cancer cells is still not well understood. We do not know whether SRPK1 is ubiquitously required for VEGF-A165 splicing, what controls SRPK1 activity, what other components of the splice machinery are required, and whether SRPK1 inhibition is necessary or sufficient for VEGF-mediated angiogenesis or other cancer processes. It has also been shown that other splice factors can control VEGF expression. In lung cancers, SRSF2 has been implicated in controlling VEGF-A165b expression (Merdzhanova et al., 2010), and in both podocytes (epithelial cells of the renal glomerulus), retinal pigmented epithelial cells (Nowak et al., 2008), and keratinocytes (Manetti et al., 2011), VEGF-A165b expression is switched on by TGFβ1-mediated activation of SRSF6. This results in overexpression of VEGF-A165b in conditions such as systemic sclerosis, where TGFβ1 is overexpressed, and inhibition of angiogenesis, resulting in ischemia and pain (Manetti et al., 2013).
VEGF-A acts through 2 cognate receptors, VEGFR1 and VEGFR2. Both of these are single transmembrane domain receptor tyrosine kinases, which can be generated by alternative splicing to form either a full-length functional protein that can bind its ligand, dimerize (hetero and homo) and signal or can form alternate splice variants that are truncated before the membrane spanning region by intron retention of intron 13 for VEGFR1 (Kendall and Thomas, 1993) or intron 10 for VEGFR2 (Vorlova et al., 2011). These splice variants result in a soluble, secreted form of the protein that acts as an antagonist to the ligands, binding them competitively with the full-length receptor and preventing angiogenesis. The control of VEGFR2 splicing is unknown, and VEGFR1 alternative splicing is still poorly understood, despite the soluble VEGFR1 isoform being pathologically upregulated in pre-eclampsia (Maynard et al., 2003), a disease that is characterized by a syndrome of increased blood pressure, reduced angiogenesis, and increased vascular permeability. Mechanisms to regulate VEGFR1 splicing have not yet been translated into therapeutics but there is potential to do so.
C. Cancer
Alternative splicing in cancer has been identified as a key contributor to the diversity of gene expression and a potential driver of nongenetic clonal expansion and epigenetic diversity (Oltean and Bates, 2014). It also suggests that resistance to treatment may be underpinned by alternative splicing and that the response of the tumor to therapy may depend on alternative splicing interactions (Abou Faycal et al., 2016). The first indication that alternative splicing was a key regulator of tumor growth was the discovery of the oncogenic nature of SRSF1 overexpression. In 2007, Adrian Krainer’s laboratory (Karni et al., 2007) at Cold Spring Harbor, demonstrated that SRSF1 underwent gene duplication in many tumors and that relatively low levels of overexpression were sufficient to transform rat fibroblasts into sarcomas. They originally showed that SRSF1 could exert these actions through three specific RNA splicing events (switching MKNK2, Bin1 and ribosomal S6 kinase 1, S6K1) but have recently identified, using an unbiased RNASeq approach, over 300 different alternatively spliced events resulting from SRSF1 overexpression in breast cancer cell lines, of which 108 were also picked up by public database approaches, and they investigated an additional five in cells in culture to show activity (Anczukow et al., 2015). The variety of cellular activity stimulated by SRSF1 includes angiogenesis (Amin et al., 2011), apoptosis (Wang et al., 2014), loss of tumor suppressor activity (Fregoso et al., 2013), translational control, and further splicing control defects (Amin et al., 2011). Interestingly they did not see activity when other SR proteins were overexpressed and, in fact, saw repression when overexpressing SRSF2 and SRSF6. These results, taken together with what is known of the effect of VEGF splicing, indicate that coordination of alternative splicing in cancer can lead to a procancer phenotype or an anticancer phenotype, depending on the activation of the different SR proteins.
Recent discoveries shed light on how primary oncogenic proteins can regulate genome-wide alternative splicing events in cancer. DNA damage appears to result in arrest of the RNA polymerase at lesions induced by, for instance, ultraviolet irradiation (Tresini et al., 2015). This results in displacement of a subset of splicing factors from the RNA polymerase and formation of RNA/DNA hybrids between the pre-mRNA and the uncoiled DNA known as R-loops (Aguilera and Garcia-Muse, 2012). This appears to be able to activate ataxia telangiectasia mutated kinase, which is now thought to be a key kinase involved in stimulating a positive feedback loop resulting in extended spliceosome mobilization, as well as a known oncogene involved in modulating DNA repair processes. Another key cancer driver, the oncogene Myc, was recently shown to drive widespread changes in splicing, including intron retention and weak 5′ donor sites, and that it can do so through interaction with specific mechanisms such as arginine methyltransferases (e.g., prmt5) (Koh et al., 2015) and BUD31 (Hsu et al., 2015), which have global effects on oncogenesis, proliferation, and apoptosis. These recent findings lend weight to the principle that targeting splicing could be a key driver for novel cancer therapeutics.
D. Pain
Mammalian pain pathways are highly complex. The peripheral signals indicative of tissue damage or infection are detected by peripheral neurons, and the input from these is processed at multiple levels of the central nervous system, from spinal cord to cortex. The ultimate perception of pain is dependent on the extent and character of this neuronal processing (Millan, 1999). As a result of this complexity, there are many cellular molecules/processes that can affect neuronal properties and therefore alter the information encoded within the pathway. Many of these molecules are known to be alternatively spliced and could therefore be potential targets for control of information transfer in the pain pathways. To date, however, very little is known about the functional implications of expression of these splice variants and much less is known about the control of the splicing. Most information is available on the functional implications of expression of different splice variants of the voltage gated calcium channels, particularly Cav2.2. An alternative splice variant of this channel is enriched in primary afferent nociceptors; the presence of this variant increases sensitivity to neuronal inhibition through opioid and GABA receptors (Bell et al., 2004; Andrade et al., 2010). Pharmacological inhibitors of this channel are in clinical usage, albeit administered intrathecally because of blood-brain barrier penetration problems, so consideration of the splice variant expression and distribution is vital if similar drugs are to be developed (McGivern, 2007). Splicing control in pain pathways is therefore an area ripe for investigation and for the development of interventions for control of pain. We recently considered the potential implications of splicing of multiple receptors/channels important in pain pathways (Donaldson and Beazley-Long, 2016), but given the early stage of this area, here we concentrate on a review of the pharmacological control of growth factor splicing in pain.
E. Growth Factors
Despite the importance of nerve growth factor (NGF) in inflammation and nociception and the existence of known splice variants, there is nothing reported on whether NGF splice variants have different physiologic actions. The splice variants of the high-affinity NGF receptor TrkA do not differ functionally. Brain-derived growth factor has multiple splice variants that, unusually, all produce the same peptide. It is thought that inclusion of specific exons in the 5′-untranslated region of RNA controls the expression of specific mRNA variants in specific tissues/conditions (Zheng et al., 2012). VEGF and VEGF receptors have only recently been implicated in nociception; inhibition of VEGF actions at VEGFR2 can result in pain (Verheyen et al., 2012; Hulse et al., 2014) and neuropathy (Verheyen et al., 2012) through a reduction of neuroprotective actions of VEGF (Verheyen et al., 2012, Beazley-Long et al., 2013). More recent evidence also shows that alternative splicing can generate algesic and analgesic VEGF isoforms, suggesting that modulation of pain by VEGF may not be solely due to actions on neuronal integrity but a direct effect of different VEGF splice variants on peripheral nociceptive neurons (Hulse et al., 2014). This is supported by the observation that alternatively spliced VEGF isoforms can be neuroprotective against neuropathy and analgesic (Hulse et al., 2015).
F. Muscle Function
Two diseases in which splicing defects have long been identified to play a key role are spinal muscular atrophy (SMA) and Duchenne’s Muscular Dystrophy (DMD). In spinal muscular atrophy there is a loss of the SMN1 gene. The homolog SMN2 can replace many of the functions of SMN1, but under normal conditions exon 7 is skipped, resulting in a premature stop codon and nonfunctional protein. Repression of exon skipping can result in a functional SMN2 protein, which can substitute for the inactive genetically deficient SMN1 gene that results in SMA (Miyaso et al., 2003). In DMD, mutations in the 3685-amino acid-long coding region that result in premature termination or nonfunctional protein, either due to mis-splicing or due to introduction of premature stop codons, result in disruption to the link between the cytoskeleton and the cell membrane. The C-terminal part of the protein is critical to its function, but the multiple rod domains in the middle of this exceptionally long protein can be missed out often with little impairment of function. Therefore a mutation in the middle of the gene that results in premature termination can often be overcome by skipping the exon containing the mutation, resulting in production of a slightly shorter protein that is nevertheless functional.
Several SFs have been implicated in the regulation of myogenesis and muscle function. Rbfox2 has been shown to coordinate a network of AS events that define myoblast differentiation and mature muscle formation. In particular, two of the Rbfox2 targets, Mef2d and Rock2, have been shown to be essential, because expression of their muscle-specific isoforms in RbFox2 depleted myoblasts rescues the impaired differentiation phenotype (Singh et al., 2014). RbFox1 is important in regulating AS in skeletal muscle but also in cardiac hypertrophy and heart failure (Pedrotti et al., 2015; Guo et al., 2012). RBM20 is a key regulator of AS in cardiac muscle and it is mutated in cardiomyopathies (Guo et al., 2012). RBM24 is an RNA-binding protein also shown to be essential in coordinating AS both in cardiac and skeletal muscle; knockout of RBM24 in mice revealed defects in heart development and sarcomerogenesis (Yang et al., 2014). MBNL and cytidine-uridine-guanidine binding protein and Elav-like family (CELF) proteins are essential in coordinating AS in heart development (Kalsotra et al., 2008) but have also been shown to be involved in myotonic dystrophy. MBNL-1 is bound by the specific expanded repeats in the 3′ untranslated region of the DMPK gene (Ho et al., 2004; Orengo et al., 2008), and its reduced availability results in defects of AS patterns and contribution to myotonic dystrophy pathogenesis.
VI. Modulators of Alternative Splicing
A. Oligonucleotides
The generation of nucleotides that can modify splicing events is now proceeding apace, from in vitro models, to in vivo models, and now into clinical trials. One approach was to take a series of SRSF1 consensus sequences followed by a sequence complementary to a specific splice site. In this case, transfection of cells with this targeted oligonucleotide enhancer of splicing results in stimulation of alternative splicing to a specific exon. This has been demonstrated in vitro in cell free systems and in intact cells, resulting in the extra inclusion of exon 7 in the SMN2 gene (Smith et al., 2014). The equivalent has also been undertaken with antisense oligonucleotides directed against SMN2 that contains the sequence for the ISS in exon 7, which then sequesters the splicing machinery that causes exon skipping, resulting in exon inclusion and rescue of the disease phenotype (Hua et al., 2007). This has now reached phase II clinical trial for SMA. The challenge with this approach is to develop compounds that can be given systemically, and that can target the appropriate cell type.
There have therefore been numerous attempts to develop therapeutic approaches for DMD using of antisense oligonucleotides, which appear to be the most advanced. Over 70% of DMD patients have mutations between exons 45 and 55 contain, so targeting this hotspot by inducing exon skipping has been a common approach. Either single exon skipping (Spitali et al., 2009; Yokota et al., 2012) or multiple exon skipping (Aartsma-Rus and van Ommen, 2009) approaches have been used using antisense sequences complementary to exon splicing enhancers. Initial phase II data on the systemic use of phosphorodiamidate morpholino oligomers, such as etiplirsen (NCT01396239) or drisapersen (NCT01153932), showed significant benefit in small cohorts of children with the disease, with the treatment preventing the decline in walking distance over a six-month trial (Mendell et al., 2013; Voit et al., 2014). These exon 51-skipping morpholino oligomers recently entered phase III (NCT02255552, NCT01480245) clinical trials, although drisapersen recently received a setback when the Food and Drug Administration concluded that the standard of substantial evidence of effectiveness had not been met.
B. RNA Binding Inhibitors/Activators
The outcome of AS events may be modulated by small molecules that bind directly to RNA, either at splice sites or at regulatory sequences (ESEs, ESSs, ISEs, ISSs). They may interfere directly with the tertiary RNA structure or hamper protein-RNA interactions. They may inhibit or activate the usage of a certain splice site. One approach has been to screen compounds for their ability to affect splicing using minigenes that express specific splice isoforms, then only selecting compounds that have specificity for that splice event (Naryshkin et al., 2014). This process has been shown to successfully identify compounds that are speculated to bind specifically to the RNA of the SMN2 gene and promote inclusion of exon 7 (Naryshkin et al., 2014). The exact nature of the interaction of this compound is not yet known, but it has been speculated that it binds either to the RNA itself or to the RNA binding domain of the splicing factors required for SMN2 exon 7 skipping (Palacino et al., 2015). One of these compounds (NVS-SM1, now known as LMI070) has now reached clinical trial (NCT02268552).
C. Splicing Factor Kinase Inhibitors
The activity of SR proteins is regulated by phosphorylation of their RS domain. Phosphorylation promotes the spliceosome assembly by facilitating protein-protein interaction; moreover it prevents the SR proteins from binding randomly on the RNA. Phosphorylation also has a role in the SR proteins’ localization and distribution in the nucleus and in the organization of the speckles, which are subnuclear structures where the SR proteins are concentrated. The phosphorylation of these proteins must be tightly regulated, because both hyper- and hypophosphorylation have been shown to reduce their overall activity (Prasad et al., 1999). Once the spliceosome is formed, dephosphorylation of the SR proteins is necessary for the transesterification cut-and-paste reaction to take place. Therefore, a cycle of phosphorylation-dephosphorylation is essential for the splicing to occur, marking the transition between stages in each round of the splicing reaction (Hagiwara, 2005; Plocinik et al., 2011). The SR protein phosphorylation levels are controlled by three main families of splicing kinases: CDC2-like kinases (CLKs), dual-specificity tyrosine-regulated kinases (DYRKs), and SR-rich splicing factor protein kinases (SRPKs). The PIM1, PIM2, and PRP4 kinases are also known to play roles in the control of phosphorylation of splicing factor proteins.
1. CDC2-like Kinases.
The CDC2-like kinases (CLKs) are an evolutionarily conserved group of dual-specificity kinases, capable of phosphorylating protein substrates on serine, threonine, and tyrosine residues. They specifically interact with RNA binding proteins, especially the members of the SR family of splicing factors (Prasad et al., 1999). CLKs contain a C-terminal domain, which is highly conserved in all the members of the family, and an N-terminal domain, which is noncatalytic and enriched with RS dipeptide residues. This region is important for interaction with SR proteins, and the main differences between family members can be found in this domain (Hanes et al., 1994; Nayler et al., 1997). The family consists of four isoforms (CLK1–4).
CLK1 (which is also known as Clk/Sty) efficiently phosphorylates SR protein SRSF1 on its RS domain, affecting its ability to bind RNA and other proteins (Colwill et al., 1996; Xiao and Manley, 1997). Catalytically inactive forms of CLK1 colocalize with SR proteins in the nuclear speckles, whereas the active form phosphorylates SR proteins, which causes speckles to dissolve and leads to the redistribution of SR proteins in the nucleus (Colwill et al., 1996). CLK1 also autophosphorylates on both serine/threonine and threonine residues, which influences the pattern of phosphorylation on SRSF1 and the ability of CLK1 to recognize different SR proteins (Prasad et al., 1999). Differential phosphorylation of SR proteins by the CLK kinases may be one of the mechanisms that characterize the late phase of virus replication. In fact, CLKs have been shown to play a role in the control of the alternative splicing of adenovirus E1A pre-mRNA in vivo (Duncan et al., 1998). CLKs have also been associated with the alternative splicing of important proteins such as tissue factor (TF) and tau. TF is an essential cofactor for the activation of blood coagulation in vivo, and elevated intravascular levels of TF have been reported in a variety of prothrombotic diseases. The pre-mRNA splicing of TF is controlled by CLK1 and is associated with increased TF protein expression, procoagulant activity, and accelerated clot formation. Interruption of CLK1 signaling prevents TF from accumulating in activated platelets (Schwertz et al., 2006). The CLKs also regulate the alternative splicing of microtubule-associated protein tau. Mutations of this protein due to aberrant splicing are implicated in frontotemporal dementia and Parkinson’s disease (Hartmann et al., 2001).
One of the first reported inhibitors of the CLK family was 5,6-dichloro-1-β-d-ribo-furanosylbenzimidazole (Fig. 3A; Nayler et al., 1998). However, this compound is also a potent inhibitor of casein kinase 2 and positive transcription elongation factor in a competitive fashion, which limits its utility (Marshall et al., 1996). Hagiwara and colleagues (Muraki et al., 2004) reported TG003 (Fig. 3B) as a small molecule with low-nanomolar IC50 values for CLK1 and CLK4. This molecule was identified from a high-throughput chemical screen of over 100,000 compounds and has proved to be a useful tool molecule. Further work by Mott et al. (2009) demonstrated that TG003 binds to CLK1 (19 nM), CLK2 (95 nM), CLK4 (30 nM), and DYRK1A (12 nM), as well as having cross reactivity with casein kinase (CK1δ and CK1ε), dual-specificity tyrosine phosphorylation-regulated kinase (DYRK1B), yeast Sps1/Ste20-related kinase (YSK4), and proviral insertion site in Moloney murine leukemia virus (PIM) kinase isoforms.
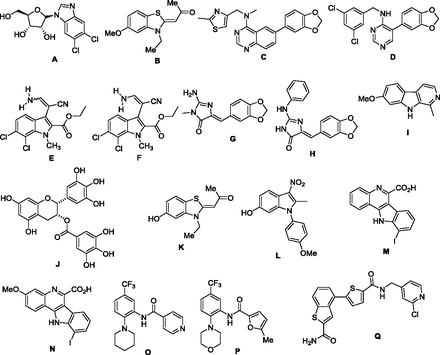
Structures of splicing factor kinase inhibitors. (A) 5,6-dichloro-1-β-d-ribo-furanosylbenzimidazole (CLK inhibitor). (B) TG003 (CLK inhibitor). (C) 6-arylquinazolin-4-amine CLK/DYRK inhibitor. (D) ML315 CLK/DYRK inhibitor. (E) KH-CB19 and (F) KH-CB20, both baurine analogs. (G) Leucettamine B. (H) Leucettine L41. (I) harmine. (J) EPCGg, (K) INDY (L) ID-8, (M and N) 10-iodo-11H-indolo[3,2-c]quinoline-6-carboxylic acids—all DYRK inhibitors. (O) SRPIN340 SRPK inhibitor. (P) SPHINX, SRPK1 inhibitor. (Q) Compound A-PRP4 inhibitor.
Thomas and coworkers (Mott et al., 2009) prepared a series of substituted 6-arylquinazolin-4-amines that were found to be potent inhibitors of the CLKs. Like TG003, it was found that these compounds also were potent DYRK1A inhibitors. The best hit (Fig. 3C) presented nanomolar activity against CLK1 (30 nM), CLK4 (50 nM), and DYRK1A (27 nM) and showed excellent selectivity when tested on a panel of 402 kinases, especially in comparison with TG003 (Mott et al., 2009; Ghosh and Adams, 2011; Coombs et al., 2013; Morooka et al., 2015). Expansion of this work was reported in 2011, with the development of 6-arylquinazolin-4-amines that have impressive potency and selectivity. Selected inhibitors possess activity versus CLK1, CLK4, and DYRK1A below 100 nM (Rosenthal et al., 2011).
Further work by Thomas, in collaboration with Aubé, has led to the report of a chemical probe ML315 (Fig. 3D), which is an aryl-substituted aminopyrimidine with activity against the CLK and DYRK families of kinases (Coombs et al., 2013). Its off-target pharmacology and in vitro pharmacokinetic properties have been further evaluated and, given that it is a selective CLK/DYRK inhibitor with adequate solubility, stability, and cell permeability, it is recognized as being suitable for cell-based biologic studies (Coombs et al., 2013).
In 2011, Knapp and coworkers (Fedorov et al., 2011) reported two new CLK1 nanomolar inhibitors, KH-CB19 (Fig. 3E, as E isomer) and KH-CB20 (Fig. 3F, as E/Z mixture), which are analogs of the marine natural product bauerine C. Both were potent CLK1 inhibitors (20 nM, 16 nM, respectively) but also had some potency against DYRK1A (55 nM, 58 nM, respectively). X-ray cocrystal structures with both CLK1 and CLK3 revealed that KHCB19 has a unique noncanonical binding mode, which helps to rationalize its selectivity.
Studies into the marine sponge natural product leucettamine B (Fig. 3G) have led to the report of a new family of inhibitors, known as leucettines, with one of them, leucettine L41 (Fig. 3H) found to inhibit CLK1 (15 nM), DYRK1A (40 nM), and DYRK2 (35 nm) (Debdab et al., 2011).
Leucettine L41 (Fig. 3H) was cocrystallized with CLK3, which confirmed that these compounds are ATP-competitive inhibitors. Importantly, G was found to inhibit the phosphorylation of SR proteins SRSF6 and SRSF4 in vitro and in vivo and was demonstrated to modulate alternative pre-mRNA splicing of Clk-1 in a cell-based reporting system (Debdab et al., 2011). The submicromolar cellular activity of these compounds makes them valuable as chemical probes.
2. Dual-specificity Tyrosine-Phosphorylation-Regulated Kinases.
The dual-specificity tyrosine-regulated kinases (DYRK) family consists of five mammalian subtypes including 1A, 1B, 2, 3, and 4. This family phosphorylates proteins on serine, threonine, and tyrosine residues and is highly conserved across species, It shows very little homology with other kinases outside the catalytic domain (Smith et al., 2012).
DYRK1A has been shown to accumulate in nuclear speckles and is known to phosphorylate a multitude of proteins involved in a wide variety of signaling pathways in vitro (Alvarez et al., 2003). In particular, DYRK1A phosphorylates the SR proteins and SF3b1, a subunit of the U2 snRNP-associated complex SF3b, the only non-SR component of the spliceosome known to be phosphorylated during splicing catalysis (Tejedor and Hämmerle, 2011; Wang et al., 1998). However, the specificity of DYRK1A in vivo is poorly defined (Tejedor and Hämmerle, 2011). Although the exact cellular functions of this kinase are still unknown, it is thought to play a critical role in the development of Down syndrome and Alzheimer’s disease. In fact, the DYRK1A gene is located within the Down syndrome critical region of chromosome 21, and increased activity of DYRK1A has been reported in various brain compartments in subjects that suffer from Down syndrome and other neurodegenerative diseases, including Alzheimer’s, Parkinson’s, Huntington’s, and Pick’s diseases (Tejedor and Hämmerle, 2011; Smith et al., 2012).
One of the first inhibitors of DYRK1A to be discovered was harmine (I, Fig. 3). It is a β-carboline alkaloid, first isolated from a South American vine, and has been shown to inhibit DYRK1A in vitro with an IC50 of 80 nM (Bain et al., 2007). However, its selectivity is poor, because it is a potent inhibitor of other important enzymes such as monoamine oxidase and cyclin-dependent kinases. Moreover it possesses significant drawbacks, such as hallucinogenic properties, due to its affinity for serotonin and tryptamine receptor binding sites. Another potent inhibitor of DYRK1A is epigallocatechin gallate (J, Fig. 3), a natural polyphenol component of green tea, with an IC50 of 330 nM (Bain et al., 2003). Although polyphenols are known to have a rather nonselective activity against a variety of enzymes and transcription factors, epigallocatechin gallate shows selective inhibition of DYRK1A and it is considered safe for human consumption, although it is not very potent.
INDY (Fig. 3K) was also developed by the Hagiwara laboratories (Ogawa et al., 2010). Although it is closely related to TG003, it has a different kinase inhibition profile. It inhibits both DYRK1A (240 nM) and DYRK1B (230 nM), while also having significant activity against DYRK2 (97% inhibition at 10 µM), CLK1 (99% at 10 µM), and CLK4 (100% at 10 µM).
ID-8 (L, Fig. 3) is an indole derivative identified by Miyabayashi et al. (2008) (as part of an investigation focused on identifying small molecules that can self-renew embryonic stem cells). Using affinity chromatography, it was found that ID-8’s molecular target was the DYRKs, although no kinase inhibition assays were carried out (Hasegawa et al., 2012).
As mentioned, many inhibitors that are active against CLKs also show potent activity on DYRK kinases, such as the 6-arylquinazolin-4-amines (C, DYRK1A 12 nM, DYRK1B 25 nM) (Rosenthal et al., 2011; Mott et al., 2009), ML315 (D, DYRK1A 282 nM) (Coombs et al., 2013), TG003 (B DYRK1A 930 nM) (Muraki et al., 2004), INDY (K DYRK1A 240 nM, DYRK1B 230 nM) (Ogawa et al., 2010), and leucettine L41 (H, DYRK1A 60 nM, DYRK1B 44 nM, DYRK2 73 nM) (Debdab et al., 2011).
Although CLKs and DYRKs are both members of the CMGC branch of the kinome that include cyclin-dependent kinases, mitogen-activated protein kinases, glycogen synthase kinase, and casein kinase 2, there is only 32.8% homology between DYRK1A and CLK1A, which would suggest that dual inhibition of these kinases should be challenging. However, closer examination of the ATP binding domain reveals that there are several conserved amino acids residues, making the ATP binding site very similar and explaining the dual inhibition observed. Until 2015, it was felt that it may not be possible to generate selective inhibitors that target just one of these kinase families. However, in 2015, Kunick and coworkers (Falke et al., 2015) reported two 10-iodo-11H-indolo[3,2-c]quinoline-6-carboxylic acids (M and N), which are the first DYRK1A-selective compounds. These two compounds have nanomolar potency against DYRK1A (6 and 22 nM, respectively). Moreover, they are selective not only versus the CLK family (for CLK1, 500 and 2000 nM respectively) but also versus the structurally and functionally related DYRK isoforms [DYRK1B (600 and >10,000 nM) and DYRK2 (>10,000 and >10,000 nM, respectively)], proving that it is possible to achieve selectivity between those kinases. Although the physicochemical properties of these compounds need to be optimized for cellular studies, the identification of this scaffold represents a significant step forward in the development of molecules that can selectively control the DYRK kinases.
3. SR Protein-specific Kinases.
SRPKs are a family of serine/arginine protein kinases that specifically phosphorylate serine in serine-arginine dipeptide residues (Papoutsopoulou et al., 1999). The human genome includes over 100 proteins containing RS domains, indicating that SRPKs could regulate many cellular functions through the phosphorylation of these substrates. A large portion of the RS domain-containing proteins is represented by the SR splicing factors, and it has been shown that these proteins are substrates for SRPKs, indicating that these kinases play a central role in the regulation of splicing. In fact, they provide a basal level of phosphorylation that is thought to play a role in the regulation of the intracellular localization of splicing factors (Giannakouros et al., 2011). The SRPK family contains three isoforms: SRPK1, SRPK2, and SRPK3. The domain arrangement in this family of kinases is unique; in fact they contain a large insert, called the spacer domain, that bifurcates the kinase core. The space domain plays a key role in the subcellular localization of these kinases (Lukasiewicz et al., 2007).
The first kinase of this family to be fully characterized was SRPK1. It consists of an amino-terminal glycine-rich region, followed by a SH3 domain (a small protein domain of about 60 amino acids residues) and a kinase domain similar to both tyrosine and serine/threonine kinases. Next to this, are two leucine/isoleucine zipper motifs and a stretch of basic amino acids. The C terminus is basic, with 24% of the residues being prolines (Gallo et al., 1994). The two ends of the previously mentioned space domain fold into unique helical structures and interact with the kinase core (Lukasiewicz et al., 2007).
SPRK1 has been shown to specifically induce the disassembly of nuclear speckles and is thought to have a central role in the regulatory network for splicing, controlling the intranuclear distribution of splicing factors in cells during interphase and the reorganization of nuclear speckles during mitosis (Colwill et al., 1996). SPRK1 also plays a critical role in regulating the function of vascular endothelial growth factor (VEGF), which, when dysregulated, stimulates blood vessel growth in cancer (Nowak et al., 2010) and induces cisplatin resistance in human ovarian carcinoma cell line (Wang et al., 2015).
SPRK2 is highly homologous to SPRK1 with the key difference being a proline-rich tract at the N terminus and an acidic region in the spacer domain (Giannakouros et al., 2011). SPRK2 specifically interacts with one of the SR proteins, acinus, which is a proapoptotic arginine-serine domain-containing protein. It has been shown that overexpression of either acinus or SRPK2 increases leukemia cell proliferation (Jang et al., 2008). Moreover both SPRK1 and SPRK2 are responsible for hepatitis B virus core protein phosphorylation during viral infection (Daub et al., 2002). SRPK3 is expressed in the heart and skeletal muscles and is required for normal muscle growth and homeostasis (Giannakouros et al., 2011).
There have been a number of SRPK inhibitors developed, including tricyclic quinoxalones (Szekelyhidi et al., 2005), with IC50 values ranging from 40 nM to 1 µM and some selectivity over other kinases (although the most potent are also c-src inhibitors), the isonicotinamides [e.g., SRPIN340 (Fig. 3O) (Fukuhara et al., 2006)] that have good selectivity and reasonable potency (∼1 µM), and the trifluoroanilino-disubstituted furans [SPHINXes (Fig. 3P) (Gammons et al., 2013b)] that are potent (<10 nM) and selective. SRPIN340 was identified via high-throughput screening and found to have low micromolar inhibition activity on SRPK1 (0.89 µM) and millimolar activity on SRPK2 (Fukuhara et al., 2006). SRPIN340 is a isonicotinamide compound that also features as key elements a phenyl ring functionalized with a trifluoromethyl group and a piperidine ring. Recent work by Morooka et al. (2015) has led to the cocrystallization of SRPIN340 with SRPK1 and revealed a unique binding mode for this kinase, although the compound is still an ATP-competitive inhibitor. The kinase catalytic domain of SRPK family members is characterized by a unique domain insert in the kinase hinge region (Ghosh and Adams, 2011). Structures of SRPK kinases revealed that a helix in this SRPK insertion domain packs adjacent to the kinase hinge region, creating a unique hydrophobic pocket located between helix αG and αH. The CF3 group on N was oriented toward this hydrophobic pocket in the unique helical insert, and to accommodate this, the backbone carbonyl of the hinge residue L168 flipped to make room for the trifluoromethyl group.
Bates, Knapp, and coworkers (Gammons et al., 2013b) used an in vitro kinase assay, as well as temperature shift assays, to identify the disubstituted furan SPHINX (P, Fig. 3) as a selective inhibitor for SPRK1 (0.44 µM). The SPHINX structure shows similarity with SRPIN340, retaining the trifluoroanilido moiety but with structural variation in the other substituents. It was found that SPHINX reduced expression of proangiogenic but not antiangiogenic VEGF isoforms. Significantly, it and SRPIN340 significantly reduced choroidal neovascularization in vivo, and as such, these molecules represent a major stepping stone for the treatment of diseases such as aged macular degeneration.
4. Pre-mRNA Processing Factor Kinase 4.
Pre-mRNA processing factor (PRP) 4 is a kinase of the DYRK family and is a serine-threonine kinase with an N-terminal arginine/serine-rich domain, and it is known for its role in regulating pre-mRNA splicing (Schneider et al., 2010). It binds pre-mRNA splicing factors SFRS8, PRP6, and pinin. It is an essential kinase that associates with both the U5 snRNP and the N-CoR deacetylase complexes. Gao et al. (2013) demonstrated how the activity of kinase PRP4 is essential for the regulation of cancer cell growth and survival, indicating the potential interest of PRP4 as a therapeutic target in oncology. The substrates for this enzyme are numerous, including other kinases and transcription factors. In this work they also reported the only inhibitor of PRP4, compound A (Q), which showed nanomolar inhibition on PRP4 (16 nM) but, unfortunately, its low cellular permeability and poor solubility will limit its application as a chemical probe.
5. Proviral insertion site of Moloney murine leukemia virus kinases.
Proviral insertion site of Moloney murine leukemia virus kinases (PIM-1, PIM-2, and PIM-3) are constitutively active serine/threonine kinases that are known to regulate cell survival (Morwick, 2010). They are overexpressed in a number of human malignancies and inflammatory states, and as such, there has been much interest in developing inhibitors of these kinases for the treatment of cancers and autoimmune diseases (Burger et al., 2013). The role of the PIM kinases in alternative splicing is not well understood, with the phosphorylation target proteins for Pim-1 and the mechanisms underlying these processes still being elucidated. However, there has been some focus on PAP-1, because this protein is implicated in autosomal dominant retinitis pigmentosa, which is the most common form of hereditary retinal degeneration. Ariga and coworkers (Maita et al., 2000, 2004) have shown that phosphorylation of PAP-1 is regulated through a Pim-1-controlled signaling pathway and that the degree of phosphorylation of PAP-1 is correlated with its splicing activity. However, to date, no reports on the use of a PIM-1 kinase inhibitor has been reported on this pathway.
VII. Considerations for Drug Development
A. Screening Cascades
To develop compounds that are able to result in altered splicing that results in improved outcome for patients, it has become necessary to set up screening cascades for therapeutic development that have some unique components to them. One example is the development of compounds that result in inhibition of splicing factor kinases, but similar approaches could be, and have been, applied to many approaches where splicing is the target. During normal drug development, a hit to lead screening process is undertaken whereby the molecular target is a key component; for instance, developing an inhibitor of the VEGFR would require a screen of a library against an in vitro kinase assay, using a high-throughput screen. For alternative splicing, the key point is whether the alternative splicing is modulated, and the target may not be identified early. So one way to screen is to undertake a high-throughput screen using a readout of splicing. Successful examples of this include using reporter genes, whereby two different colors are generated according to the splice site (Bonano et al., 2007). These minigenes have been developed for a while against alternatively spliced proteins such as SMN2 (Cherry et al., 2012) and FGFRIII (Bonano et al., 2006). A high-throughput screen can then be undertaken using cells as a marker to identify compounds that can affect splicing of the specific gene (Cherry et al., 2012). However, this approach is likely to result in a large number of false positives, because compounds that differentially affect transcription could also alter splicing because of the processivity of the spliceosome—increased (or decreased) transcription rates can change the splice sites identified (Nogues et al., 2003). Therefore using multiple reporters to identify that the gene of interest is alternatively spliced and other alternatively spliced genes are not altered is useful. Second, the screening cascade needs to ensure that the effect is specific, so testing the effect across a variety of spliced products that are known to be within the target tissue (i.e., within muscle, are there effects on other known splice variants?) or within the known target profile (i.e., for SRSF1 mediated targets, are all SRSF1 known genes altered or just a few?). This can also be done across the whole transcriptome using RNA sequencing followed by bioinformatic analysis that allows the difference in mRNA expression between two isoforms from the same gene to be examined in highly expressing genes. This type of specificity screening, as shown by Naryshkin et al. (2014), indicates that specificity for splicing can be achieved. A series of toxicity analyses needs to be carried out as would be the case for any other drug, but in this case it is worth considering that the effects may not be on the potential targets themselves, but on alternative splicing of the targets. For instance, many studies need to investigate the action of the drug on the human ether-a-go go-related gene product—a potassium channel involved in repolarization of the cardiac action potential. Standard screening cascades would include a hERG activity screen using cells overexpressing hERG from a cDNA. If the effect is not on the channel itself but on alternative splicing of the channel then endogenous hERG expressing cells need to be investigated instead (Kupershmidt et al., 1998). This also applies to numerous other toxicity and pharmacokinetic studies, including CYP inhibition, metabolism, and breakdown of the targets.
B. Biomarkers
For most drugs, the knowledge that the agent is having the desired biologic effect is a critical part of the development program. A biomarker of efficacy is key. However, for alternative splicing modulators this can pose a problem, because there may be multiple downstream pathways that are modified and these may be cell type, tissue type, and context dependent. For instance, targeting a component of the alternative splicing pathway such as a SR protein may produce different downstream alternative splicing (or none) in some tissues and desirable consequences in the target tissue. Thus biomarkers of efficacy need to be chosen carefully and specifically for each target type. SR kinase inhibitors can use the target phosphorylation as a biomarker of efficacy, for instance by immunoprecipitation of the SR protein followed by Western blotting for phosphoserine or phosphoSR, but inhibitors of the spliceosome such as spliceostatin have a more difficult biomarker problem, because the effect will be to disrupt splicing of many genes, and the therapeutic window is therefore narrower. Using oligo-targeted therapies gives a more subtle biomarker, because it is possible that only a single RNA species may be changed. It is difficult using standard quantitative RNA techniques to measure RNA changes in splicing quantitatively, because qPCR is designed to detect fold changes or order of magnitude changes due to the logarithmic relationship of PCR. A novel technology to determine splicing changes that has been developed is digital droplet PCR (Hindson et al., 2013), which is capable of quantifying a few percent change (Sun et al., 2014), and has been used to identify changes in RNA splice variants of androgen receptor in a few circulating tumor cells in prostate cancer (Ma et al., 2016). Alternatively RNA-seq can be used to determine splicing changes by investigating splice junction reads (Anczukow et al., 2015). This is expensive but can be highly sensitive.
C. Off Target Effects
As with any drug development, possible off-target effects should be considered. Relatively recently there has been a reluctance in the field that specificity in splicing therapeutics may be ever achieved, knowing the immense number of splice sites and the conserved consensus sequence at these sites. However, clues that this might be possible came from the elucidation of how specificity of splice sites choice is accomplished through a complex interaction of RNA tertiary structure and SFs. Indeed, two recent screens from Roche and Novartis of cells expressing splicing reporters (HEK cells or NSC34 motor neuron cell lines, respectively) using large chemical libraries (Naryshkin et al., 2014; Palacino et al., 2015) have revealed small molecules that are incredibly specific for a certain splice site targeted in the SMN gene. RNA-seq analysis has shown a very small number of additional splice sites being affected.
Targeting splicing regulators that are once or twice removed from RNA (e.g., splicing factor kinases) is likely to affect several splice sites and genes. However, alternative splicing is cell and tissue specific, and therefore it will be important to define functionally whether modification of additional splice sites than those intended is important in the system to be studied. Indeed, we recently showed that knockdown of SRPK1 in PC3 prostate cancer cells predominantly affects their angiogenesis potential through modifying VEGF splicing (Mavrou et al., 2015) and not other properties (e.g., proliferation, migration), whereas in breast cancer cell lines SRPK1 knockdown induces apoptosis (Hayes et al., 2007), but the mechanisms behind this specificity are not yet elucidated.
Off target effects could also be limited by designing routes of administration of inhibitors—for instance using topical delivery of compounds or by injection into a restricted space such as the vitreous of the eye. This approach has been used in preclinical models for SRPK1 inhibitors in models of retinopathy of prematurity (Gammons et al., 2013a) and choroidal neovascularization (Gammons et al., 2013b).
D. Safety
The safety profile of modulators of alternative splicing is regularly questioned during the development of modulators of this process. The two different types of splicing modulation—constitutive and alternative splicing—are expected to have substantially different safety and toxicity profiles. Early results with modulators of constitutive splicing indicated substantial toxicity, but recent studies with the SF3B1 inhibitor spliceostatin and pladeolinide B indicate that even targeting constitutive splicing may not have toxicity levels greater than that associated with modulators of other constitutive cellular processes, such as the cell cycle inhibitors, cytoskeletal inhibitors or channel blockers.
The safety profile of alternative splicing modulators has not been clearly established and, of course, is likely to be different for each pathway, but preliminary studies appear to provide remarkably good safety profiles. Inhibitors of SRPK1 have been shown to be given systemically at supratherapeutic doses in animals without any clear side effects (Fukuhara et al., 2006), and nucleotide specific inhibitors are considered to be highly specific and therefore less prone to toxic side effects. The recent suspension of the clinical trial involving alternative splicing inhibitors of the SM2 gene due to potential off target effects in animal models will be an interesting development to follow, but even here, no clear indication of toxicity was reported in patients at this time.
VIII. Conclusions and Future Directions
The development of splicing inhibitors as therapeutic compounds is in its infancy, with the first targeted splicing inhibitors only just reaching phase III clinical trials. However, compounds that regulate splicing are already used in clinical practice—valproate (Brichta et al., 2006), amiloride (Chang et al., 2011), and caffeine (Shi et al., 2008)—have all been shown to regulate splicing (of SMN2, BClx, and KLF6, respectively) through modulation of splicing factors. The new generation of specific highly targeted compounds is likely to usher in a new era of splicing regulators, with multiple mechanisms of action and targets.
Authorship Contributions
Participated in research design: Bates, Morris, Oltean, and Donaldson.
Wrote or contributed to the writing of the manuscript: Bates, Morris, Oltean, and Donaldson.
Footnotes
This work was funded by the MRC [MR/N01247X/1 (D.O.B.), MR/L01985X/1, (D.O.B.) MR/K020366/1 (D.O.B., S.O.), MR/K013157/1 (D.O.B.)], BBSRC BB/J007293/1 (D.O.B.), BHF (PG/15/53/31371, S.O.), and ARUK (20400, L.F.D.).
Abbreviations
- AS
- alternative splicing
- CLK
- CDC2-like kinase
- DMM
- Duchenne’s muscular dystrophy
- DYRK
- dual-specificity tyrosine-regulated kinases
- ESE
- exonic splicing enhancer
- ESS
- exonic splicing silencer
- hnRNP
- heterogeneous nuclear ribonucleoproteins
- ISE
- intronic splicing enhancer
- ISS
- intronic splicing silencer
- NGF
- nerve growth factor
- PIM
- proviral insertion site in Moloney murine leukemia virus
- PRP
- pre-mRNA processing factor
- SF
- splice factor
- SMA
- spinal muscle atrophy
- SRPK
- SR-rich splicing factor protein kinases
- TF
- tissue factor
- VEGF
- vascular endothelial growth factor
- Copyright © 2016 by The Author(s)
This is an open access article distributed under the CC BY-NC Attribution 4.0 International license.