Abstract
The study of antiviral pathways to reveal methods for the effective response and clearance of virus is closely related to understanding interferon (IFN) signaling and its downstream target genes, IFN-stimulated genes. One of the key antiviral factors induced by IFNs, 2′-5′ oligoadenylate synthase (OAS), is a well-known molecule that regulates the early phase of viral infection by degrading viral RNA in combination with RNase L, resulting in the inhibition of viral replication. In this review, we describe OAS family proteins from a different point of view from that of previous reviews. We discuss not only RNase L-dependent (canonical) and -independent (noncanonical) pathways but also the possibility of the OAS family members as biomarkers for various diseases and clues to non-immunological functions based on recent studies. In particular, we focus on OASL, a member of the OAS family that is relatively less well understood than the other members. We will explain its anti- and pro-viral dual roles as well as the diseases related to single-nucleotide polymorphisms in the corresponding gene.
Similar content being viewed by others
Introduction
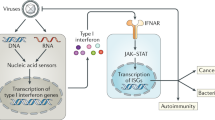
To counter virus infection, the immune system produces antiviral cytokines. Interferon (IFN) is the most powerful antiviral cytokine, and it induces IFN-stimulated genes that mediate antiviral effector functions. Among the proteins induced by IFN, the oligoadenylate synthase (OAS) proteins have been identified as enzymes that sense exogenous nucleic acid and initiate antiviral pathways. The OAS family proteins belong to a template-independent nucleotidyltransferase family. The oligomerized OAS enzyme generates the 2′-5′-linked oligoadenylate (2-5A) activating endoribonuclease, RNase L, which degrades cellular and viral RNA. In this way, RNase L contributes to the control of the early spread of a virus by degrading the viral RNA and activating cytoplasmic pattern-recognition receptors, including RIG-I and MDA-5 (Figure 1). Although the antiviral activity of the OAS family and 2-5A-RNase L is well characterized,1, 2, 3 it was reported recently that not all of the OAS antiviral function is mediated by the RNase L-dependent pathway.4, 5 This indicates that the OAS family proteins may be involved in pathways for regulating viral infection other than the pathway involving RNase L activation.
The OAS family in antiviral pathways. Following virus infection of a host cell, viral dsRNA stimulates OAS1, OAS2 and OAS3 and leads to the synthesis of 2′-5′ oligoA and RNase L activation. Activated RNase L cleaves viral and cellular RNA. MDA5 and RIG-I detect cleaved viral RNA, thereby promoting the activation of IRF3 and IRF7. The hOASL binds to RIG-I and enhances the sensitivity of RIG-I signaling through the UBL domain, whereas the mOASL1 binds to IRF7 mRNA and inhibits the translation of IRF7.
The OAS family proteins consist of OAS1, OAS2, OAS3 and OAS-like protein (OASL).6 The OAS1-3 proteins have significant homology to each other and only differ in the number of OAS units. OAS1, OAS2 and OAS3 contain one, two and three OAS units, respectively. The genes encoding the OAS proteins are clustered on chromosome 12 (in the 12q24.1 region) in humans. Human OAS1 (hOAS1) has two spliced forms that produce proteins of 40 and 46 kDa, each containing a distinct C-terminal sequence.7 Three more alternative splice forms of Oas1 are generated by single-nucleotide polymorphisms (SNPs).8 There are two isoforms of OAS2 (p69 and p71), and one 100 kDa OAS3 form.7 Two isoforms of hOASL OASLa (p59) and OASLb (p30) are coded by a gene located on chromosome 12 (in the 12q24.2 region), but OASLa is the dominant isoform; OASLb has a C-terminal truncation. Recently, another OASL isoform, OASLd, which is strongly induced by IFNγ, was discovered.9 In the mouse genome, eight Oas1 genes and one gene each for OAS2 and OAS3 exist on chromosome 8.10 Mice have two OASL genes (Oasl1 and Oasl2) on chromosome 5.11 Studies using genetically modified mouse models for the OAS genes may allow us to gain insights into the human OAS family functions, because the exon/intron structures of all the genes are conserved between humans and mice.10 In this article, we review the immune-related and other cellular functions of OAS and the dual role of OASL. We then discuss SNPs and the differential expression of OAS in various diseases.
RNase L-dependent and -independent antiviral functions of OAS1, OAS2 and OAS3
The canonical OAS/RNase L pathway has been intensively studied, and the results have been well documented in several reviews.6, 12 The ability to activate RNase L of the OAS family is dependent on the OAS oligomerization unit and the processivity of synthesizing dimeric or tri-tetrameric 2-5A. The CFK motif in the OAS domain, which is required for tetramerization of OAS and affects the synthesis of effective 2-5A, is only conserved between OAS1 and OAS2.13 Originally, it was believed that only OAS1 and OAS2, and not OAS3, contributed to RNase L-dependent antiviral activity.13, 14 This idea has been revised by studies of mouse OAS1b. The observation that point mutations of mOas1b increased the susceptibility of mice to West Nile Virus (WNV) indicated that the OAS/RNase L system is required for virus restriction.5, 15 Recent experiments with each of the isoforms of the OAS family overexpressed cell revealed that OAS3 shows RNase L-dependent antiviral activity against dengue virus.16 Furthermore, purified OAS3 can produce long enough 2-5A to activate RNase L.17 In an antiviral response to hepatitis C virus, overexpressed OAS1 p46 and OAS3 p100 show antiviral activity by mediating the RNase L-dependent pathway.18 OAS3 synthesizes dimeric 2-5A that binds to RNase L with low affinity and produces 2-5A oligomers shorter than the tri-tetramer 2-5As produced by OAS1 and OAS2. However, OAS3 can be induced by smaller amounts of double-stranded RNA (dsRNA) than OAS1 and OAS2.19 The higher dsRNA-binding affinity of OAS3 to dsRNA might compensate for the lower binding affinity of dimeric 2-5A to RNase L.
Recent studies have focused on the OAS expression patterns after various virus infections and the polymorphisms associated with virus infection susceptibility. Dengue virus infection causes the early induction of OAS1, whereas OAS2 and OAS3 are upregulated later.20, 21 Studies of polymorphisms in the Oas genes have emphasized the importance of all the three Oas genes during dengue virus infection.22 In the case of Chikungunya virus (CHIKV), OAS3 shows a strong correlation with resistance to CHIKV infection. Mutation of Oas3 caused less efficient inhibition of CHIKV, regardless of RNase L activation.23 CHIKV with the E2-E116K substitution escaped the antiviral action of OAS3 and replicated more successfully than the wild-type virus by reducing OAS3 expression.24, 25 Therefore, OAS3 appears to be required for the restriction of specific viruses, independent of RNase L.
The OASL: an antiviral or pro-viral protein?
OASL contains one OAS domain, two ubiquitin-like repeats and a CCY motif instead of CFK in the OAS unit that is required for oligomerization. Thus the hOASL lacks the 2′-5′-linked OAS activity that is one of the hallmarks of OAS.26 Expression of OASL is also regulated differently from OAS.27 Whereas mOASL1 is mainly induced by IFNa, mOASL2 is upregulated by both type I and type II IFN.28 Unlike hOASL, mOASL2 maintains OAS activity and requires dsRNA as an inducer. The amino-acid sequence of mOASL1 is more similar to human OASL than mOASL2 is (mouse OASL1, 74%; mouse OASL2, 49% identical with hOAL).10 Thus it is plausible that mOASL1 is the functional counterpart and ortholog of hOASL. It has been suggested that mOASL2 is an intermediate between OAS and OASL that arose during evolution.11
The role of OASL was identified later than that of OAS, but the recently identified functions seem to be critical to the antiviral innate and adaptive immune responses. These facts make further studies of OASL valuable. Because of its OAS-like qualities (activation by IFN and binding to dsRNA) and inactive nucleotidyltransferase domain, OASL has been assumed to interfere with the 2-5A and RNase L pathway by competing with OAS.29, 30 An SNP study of the response to IFN therapy for chronic hepatitis C suggested that OASL might negatively regulate the antiviral function of OAS. An SNP that cause lower levels of OASL was observed to be associated with a sustained virological response after treatment.31 However, it has been demonstrated that hOASL possesses antiviral activity through the C-terminal ubiquitin-like domain. In Vero cells, hOASL expression increases the resistance against single-stranded RNA viruses, including picornavirus and encephalomyocarditis, but not against a DNA virus, herpes simplex virus 1.32 A recent study using an OASL deletion mutant produced by a transcription activator-like effector nucleases (TALEN) procedure found that hOASL suppresses the replication of vesicular stomatis virus by enhancing the RIG-I pathway (Figure 1). In addition, this study established that mOASL2 also has an antiviral activity, suggesting that mOASL2 is functionally comparable to hOASL.33 The function of mOASL1 is totally different from that of mOASL2 and hOASL. The mOASL1 protein inhibits the translation of IFN-regulating transcription factor 7, the main transcription factor for type I IFN, and suppresses the production of type I IFN during viral infection34 (Figure 1) through an interaction with the stem loop structure in the 5′-untranslated region of IFN-regulating transcription factor 7.35 Negative regulation of type I IFN production by mOASL1 causes viral persistence and represses T-cell function.36 Together, OASL proteins have dual functions that depend on various mechanisms and on the phase of the virus infection.
The OAS family as a biomarker
Expression of the OAS family is upregulated in some diseases whether it is dependent or independent of IFN stimuli. In light of this, OAS can be a useful biomarker for various diseases in multiple stages. A specific biomarker is needed to select an adequate therapy for an individual and to monitor the effect of the therapy. The OAS level is strongly related to autoimmune diseases and chronic infections, including systemic lupus erythematosus, systemic sclerosis, rheumatoid arthritis and multiple sclerosis.37, 38, 39, 40 Appropriate biomarkers must express specific patterns depending on the condition of a disease. In the study of systemic lupus erythematosus, it was shown that OAS1 is upregulated, whereas expression of OASL was lower in systemic lupus erythematosus patients than in normal individuals.41 In primary human peripheral blood mononuclear cells isolated from systemic sclerosis patients, only the expression of OAS2 and OASL was higher than in the basal state and neither OAS1 nor OAS3 was induced.37 Patients with severe chronic obstructive pulmonary disease show high mortality. Exposure to influenza virus and cigarette smoke leads to a more serious stage of chronic obstructive pulmonary disease. In a mouse experiment, only OAS2 and OASL were synergistically induced by cigarette smoke and influenza virus, whereas OAS1 and OASL2 were not induced.42 OASL also can be used as a biomarker to predict rheumatoid arthritis patients’ response to tocilizumab, the drug most commonly used to treat this disease. Expression of OASL differs significantly between nonresponders and responders.43 The expression pattern of the OAS family may offer useful information for therapy of autoimmune diseases and chronic infections and reveal different roles of each member of the OAS family in autoimmune disorders.
SNPs in OASL-associated diseases
Several SNPs in the OAS family genes have been identified and associated with various diseases (Table 1). Consistent with the main role of OAS proteins, SNPs in OAS genes affect the susceptibility to viral infection. Most people who are infected with WNV of the Flaviviridae family show no symptoms, but a few people (<1%) progress to a severe clinical infection, West Nile fever.44 Genetic factors, age and environmental conditions may have essential roles in disease progression. Sequencing the OAS family exons in 33 patients with WNV infection showed that a SNP (rs3213545) of the OASL gene is associated with WNV infection.45 Although the rs3213545 SNP is synonymous in OASL exon2, it contains a splice enhancer site that is a minor allele ‘T’, which generates a dominant-negative mutant form of OASL. A study using 331 WNV-infected patients showed that the OAS1 SNP rs10774671 has a significant link with WNV. These data support the conclusion that human OAS1 and OASL have antiviral roles against WNV. In contrast, the minor allele ‘T’, the rs3213545 SNP, is significantly associated with a sustained virological response, an efficacy measure of hepatitis C virus treatment after IFN therapy;31 that is, diminished OASL activity confers an advantage for IFN treatment. Depending on the early response to the recovery phase, one of the dual roles of OASL can be addressed.
The role of OASL in diseases other than immune-related diseases is demonstrated by genome-wide association studies. The locus of the OASL gene on chromosome 12 was shown to affect multiple cardiovascular-related traits, especially ‘high or low’ gamma glutamyltransferase, low-density lipoprotein and C-reactive protein. The study identified the rs3213545 SNP as a possible candidate associated with liver function and lipid constitution.46
Non-immunological functions of the OAS family proteins
Although the major role of the OAS proteins is as immune regulators, there are some data showing that OAS proteins are involved in other cellular functions. The OAS family is associated with the regulation of apoptosis, one of the ways organisms react against virus infection in an effort to eliminate the virus-infected cells and a core mechanism for inhibiting tumorigenesis.47, 48 A derivative of the dimeric 2-5A-activating RNase L synthesized by OAS3 has been described as a new molecule for inhibiting breast cancer cell growth.49 OAS3 is also one of the genes in a breast tumor cell line (MCF7) that is highly expressed after daily exposure to radiation.50 In addition, the OAS/RNase L pathway is induced by BRCA1, a tumor suppressor of the breast and ovarian cancer, and it activates apoptosis of tumor cells.51 These observations implicate the OAS proteins in counteracting tumor progression.
Even though hOASL has a supposed nuclear localization signal (RKVKEKIRRTR) at the C-terminus, its nuclear function is not clear.6 However, there is some evidence that a nuclear function of OASL exists. A yeast two-hybrid system identified an interaction between OASL and methyl CpG-binding protein 1, which is a transcriptional repressor.52 In addition, it has been shown that OAS might also regulate nuclear events, including pre-mRNA splicing, a complicated process that requires a complex of RNA and proteins. OAS assembles the 60S spliceosome and is necessary for the first step of splicing.53
Conclusion
The RNaseL-dependent immune regulation by the OAS family is widely known. However, various functions of each OAS family member and a newly defined mechanism independent of RNase L have been discovered recently. Because most of the studies focusing on the RNase L-independent functions of OAS employ in vitro methods, including genetic depletion, using an in vivo knockout mouse model would be useful for finding new mechanisms. Other important aspects are SNPs and the expression level changes of the OAS family during disease progression. Many SNPs in the OAS genes have been discovered to be associated with viral infections and autoimmune diseases. In addition, the expression levels of OAS family members depend on disease progression. Although we still lack a complete understanding of the roles of SNPs of the OAS family members in disease, their strong correlation with viral diseases will prove useful for identifying new treatment methods for infectious and autoimmune diseases.
References
Hovanessian AG, Wood JN . Anticellular and antiviral effects of pppA(2'p5'A)n. Virology 1980; 101: 81–90.
Chebath J, Benech P, Revel M, Vigneron M . Constitutive expression of (2'-5') oligo A synthetase confers resistance to picornavirus infection. Nature 1987; 330: 587–588.
Schroder HC, Suhadolnik RJ, Pfleiderer W, Charubala R, Muller WE . 2'-5')Oligoadenylate and intracellular immunity against retrovirus infection. Int J Biochem 1992; 24: 55–63.
Kristiansen H, Scherer CA, McVean M, Iadonato SP, Vends S, Thavachelvam K et al. Extracellular 2'-5' oligoadenylate synthetase stimulates RNase L-independent antiviral activity: a novel mechanism of virus-induced innate immunity. J Virol 2010; 84: 11898–11904.
Kajaste-Rudnitski A, Mashimo T, Frenkiel MP, Guenet JL, Lucas M, Despres P . The 2',5'-oligoadenylate synthetase 1b is a potent inhibitor of West Nile virus replication inside infected cells. J Biol Chem 2006; 281: 4624–4637.
Sadler AJ, Williams BR . Interferon-inducible antiviral effectors. Nat Rev Immunol 2008; 8: 559–568.
Hovnanian A, Rebouillat D, Mattei MG, Levy ER, Marie I, Monaco AP et al. The human 2',5'-oligoadenylate synthetase locus is composed of three distinct genes clustered on chromosome 12q24.2 encoding the 100-, 69-, and 40-kDa forms. Genomics 1998; 52: 267–277.
Bonnevie-Nielsen V, Field LL, Lu S, Zheng DJ, Li M, Martensen PM et al. Variation in antiviral 2',5'-oligoadenylate synthetase (2' 5' AS) enzyme activity is controlled by a single-nucleotide polymorphism at a splice-acceptor site in the OAS1 gene. Am J Hum Genet 2005; 76: 623–633.
Guo X, Li X, Xu Y, Sun T, Yang G, Wu Z et al. Identification of OASL d, a splice variant of human OASL, with antiviral activity. Int J Biochem Cell Biol 2012; 44: 1133–1138.
Eskildsen S, Hartmann R, Kjeldgaard NO, Justesen J . Gene structure of the murine 2'-5'-oligoadenylate synthetase family. Cell Mol Life Sci 2002; 59: 1212–1222.
Eskildsen S, Justesen J, Schierup MH, Hartmann R . Characterization of the 2'-5'-oligoadenylate synthetase ubiquitin-like family. Nucleic Acids Res 2003; 31: 3166–3173.
Chakrabarti A, Jha BK, Silverman RH . New insights into the role of RNase L in innate immunity. J Interferon Cytokine Res 2011; 31: 49–57.
Ghosh A, Sarkar SN, Guo WD, Bandyopadhyay S, Sen GC . Enzymatic activity of 2'-5'-oligoadenylate synthetase is impaired by specific mutations that affect oligomerization of the protein. J Biol Chem 1997; 272: 33220–33226.
Dong B, Xu L, Zhou A, Hassel BA, Lee X, Torrence PF et al. Intrinsic molecular activities of the interferon-induced 2-5A-dependent RNase. J Biol Chem 1994; 269: 14153–14158.
Courtney SC, Di H, Stockman BM, Liu H, Scherbik SV, Brinton MA . Identification of novel host cell binding partners of Oas1b, the protein conferring resistance to flavivirus-induced disease in mice. J Virol 2012; 86: 7953–7963.
Lin RJ, Yu HP, Chang BL, Tang WC, Liao CL, Lin YL . Distinct antiviral roles for human 2',5'-oligoadenylate synthetase family members against dengue virus infection. J Immunol 2009; 183: 8035–8043.
Ibsen MS, Gad HH, Thavachelvam K, Boesen T, Despres P, Hartmann R . The 2'-5' oligoadenylate synthetase 3 (OAS3) enzyme potently synthesizes the 2'-5' oligoadenylates required for RNase L activation. J Virol 2014; 88: 14222–14231.
Kwon YC, Kang JI, Hwang SB, Ahn BY . The ribonuclease L-dependent antiviral roles of human 2',5'-oligoadenylate synthetase family members against hepatitis C virus. FEBS Lett 2013; 587: 156–164.
Rebouillat D, Hovanessian AG . The human 2',5'-oligoadenylate synthetase family: interferon-induced proteins with unique enzymatic properties. J Interferon Cytokine Res 1999; 19: 295–308.
Bordignon J, Probst CM, Mosimann AL, Pavoni DP, Stella V, Buck GA et al. Expression profile of interferon stimulated genes in central nervous system of mice infected with dengue virus Type-1. Virology 2008; 377: 319–329.
Warke RV, Xhaja K, Martin KJ, Fournier MF, Shaw SK, Brizuela N et al. Dengue virus induces novel changes in gene expression of human umbilical vein endothelial cells. J Virol 2003; 77: 11822–11832.
Alagarasu K, Honap T, Damle IM, Mulay AP, Shah PS, Cecilia D . Polymorphisms in the oligoadenylate synthetase gene cluster and its association with clinical outcomes of dengue virus infection. Infect Genet Evol 2013; 14: 390–395.
Brehin AC, Casademont I, Frenkiel MP, Julier C, Sakuntabhai A, Despres P . The large form of human 2',5'-oligoadenylate synthetase (OAS3) exerts antiviral effect against Chikungunya virus. Virology 2009; 384: 216–222.
Henrik Gad H, Paulous S, Belarbi E, Diancourt L, Drosten C, Kummerer BM et al. The E2-E166K substitution restores Chikungunya virus growth in OAS3 expressing cells by acting on viral entry. Virology 2012; 434: 27–37.
Priya R, Patro IK, Parida MM . TLR3 mediated innate immune response in mice brain following infection with Chikungunya virus. Virus Res 2014; 189: 194–205.
Hovnanian A, Rebouillat D, Levy ER, Mattei MG, Hovanessian AG . The human 2',5'-oligoadenylate synthetase-like gene (OASL) encoding the interferon-induced 56-kDa protein maps to chromosome 12q24.2 in the proximity of the 2',5'-OAS locus. Genomics 1999; 56: 362–363.
Melchjorsen J, Kristiansen H, Christiansen R, Rintahaka J, Matikainen S, Paludan SR et al. Differential regulation of the OASL and OAS1 genes in response to viral infections. J Interferon Cytokine Res 2009; 29: 199–207.
Eskildsen S, Justesen J, Schierup MH, Hartmann R . Characterization of the 2'-5'-oligoadenylate synthetase ubiquitin-like family. Nucleic Acids Res 2003; 31: 3166–3173.
Rogozin IB, Aravind L, Koonin EV . Differential action of natural selection on the N and C-terminal domains of 2'-5' oligoadenylate synthetases and the potential nuclease function of the C-terminal domain. J Mol Biol 2003; 326: 1449–1461.
Harcourt JL, Hagan MK, Offermann MK . Modulation of double-stranded RNA-mediated gene induction by interferon in human umbilical vein endothelial cells. J Interferon Cytokine Res 2000; 20: 1007–1013.
Su X, Yee LJ, Im K, Rhodes SL, Tang Y, Tong X et al. Association of single nucleotide polymorphisms in interferon signaling pathway genes and interferon-stimulated genes with the response to interferon therapy for chronic hepatitis C. J Hepatol 2008; 49: 184–191.
Marques J, Anwar J, Eskildsen-Larsen S, Rebouillat D, Paludan SR, Sen G et al. The p59 oligoadenylate synthetase-like protein possesses antiviral activity that requires the C-terminal ubiquitin-like domain. J Gen Virol 2008; 89: 2767–2772.
Zhu J, Zhang Y, Ghosh A, Cuevas RA, Forero A, Dhar J et al. Antiviral activity of human OASL protein is mediated by enhancing signaling of the RIG-I RNA sensor. Immunity 2014; 40: 936–948.
Lee MS, Kim B, Oh GT, Kim YJ . OASL1 inhibits translation of the type I interferon-regulating transcription factor IRF7. Nat Immunol 2013; 14: 346–355.
Kim YM, Choi WY, Oh CM, Han GH, Kim YJ . Secondary structure of the Irf7 5'-UTR, analyzed using SHAPE (selective 2'-hydroxyl acylation analyzed by primer extension). BMB Rep 2014; 47: 558–562.
Lee MS, Park CH, Jeong YH, Kim YJ, Ha SJ . Negative regulation of type I IFN expression by OASL1 permits chronic viral infection and CD8(+) T-cell exhaustion. PLoS Pathog 2013; 9: e1003478.
Almeida GMD, de Oliveira DB, Botelho LM, Silva LKD, Guedes ACM, Santos FPST et al. Differential upregulation of human 2' 5' OAS genes on systemic sclerosis: detection of increased basal levels of OASL and OAS2 genes through a qPCR based assay. Autoimmunity 2014; 47: 119–126.
Croze E . Differential gene expression and translational approaches to identify biomarkers of interferon beta activity in multiple sclerosis. J Interferon Cytokine Res 2010; 30: 743–749.
Preble OT, Rothko K, Klippel JH, Friedman RM, Johnston MI . Interferon-induced 2'-5' adenylate synthetase in vivo and interferon production in vitro by lymphocytes from systemic lupus erythematosus patients with and without circulating interferon. J Exp Med 1983; 157: 2140–2146.
Hertzog PJ, Emery P, Cheetham BF, Mackay IR, Linnane AW . Interferons in rheumatoid arthritis: alterations in production and response related to disease activity. Clin Immunol Immunopathol 1988; 48: 192–201.
Ye S, Guo Q, Tang JP, Yang CD, Shen N, Chen SL . Could 2' 5'-oligoadenylate synthetase isoforms be biomarkers to differentiate between disease flare and infection in lupus patients? A pilot study. Clin Rheumatol 2007; 26: 186–190.
Zhou Y, Kang MJ, Jha BK, Silverman RH, Lee CG, Elias JA . Role of ribonuclease L in viral pathogen-associated molecular pattern/influenza virus and cigarette smoke-induced inflammation and remodeling. J Immunol 2013; 191: 2637–2646.
Sanayama Y, Ikeda K, Saito Y, Kagami S, Yamagata M, Furuta S et al. Prediction of therapeutic responses to tocilizumab in patients with rheumatoid arthritis biomarkers identified by analysis of gene expression in peripheral blood mononuclear cells using genome-wide DNA microarray. Arthritis Rheumatol 2014; 66: 1421–1431.
Mostashari F, Bunning ML, Kitsutani PT, Singer DA, Nash D, Cooper MJ et al. Epidemic West Nile encephalitis, New York, 1999: results of a household-based seroepidemiological survey. Lancet 2001; 358: 261–264.
Yakub I, Lillibridge KM, Moran A, Gonzalez OY, Belmont J, Gibbs RA et al. Single nucleotide polymorphisms in genes for 2'-5'-Oligoadenylate synthetase and RNase L in patients hospitalized with West Nile virus infection. J Infect Dis 2005; 192: 1741–1748.
Middelberg RPS, Ferreira MAR, Henders AK, Heath AC, Madden PAF, Montgomery GW et al. Genetic variants in LPL, OASL and TOMM40/APOE-C1-C2-C4 genes are associated with multiple cardiovascular-related traits. BMC Med Genet 2011; 12: 123.
Domingo-Gil E, Esteban M . Role of mitochondria in apoptosis induced by the 2-5A system and mechanisms involved. Apoptosis 2006; 11: 725–738.
Castelli JC, Hassel BA, Maran A, Paranjape J, Hewitt JA, Li XL et al. The role of 2'-5' oligoadenylate-activated ribonuclease L in apoptosis. Cell Death Differ 1998; 5: 313–320.
Latham KE, Cosenza S, Reichenbach NL, Mordechai E, Adelson ME, Kon N et al. Inhibition of growth of estrogen receptor positive and estrogen receptor negative breast cancer cells in culture by AA-etherA, a stable 2-5A derivative. Oncogene 1996; 12: 827–837.
Tsai MH, Cook JA, Chandramouli GV, DeGraff W, Yan H, Zhao S et al. Gene expression profiling of breast, prostate, and glioma cells following single versus fractionated doses of radiation. Cancer Res 2007; 67: 3845–3852.
Mullan PB, Hosey AM, Buckley NE, Quinn JE, Kennedy RD, Johnston PG et al. The 2,5 oligoadenylate synthetase/RNaseL pathway is a novel effector of BRCA1- and interferon-gamma-mediated apoptosis. Oncogene 2005; 24: 5492–5501.
Andersen JB, Strandbygard DJ, Hartmann R, Justesen J . Interaction between the 2'-5' oligoadenylate synthetase-like protein p59 OASL and the transcriptional repressor methyl CpG-binding protein 1. Eur J Biochem 2004; 271: 628–636.
Sperling J, Chebath J, Araddann H, Offen D, Spann P, Lehrer R et al. Possible involvement of (2'-5')oligoadenylate synthetase-activity in pre-messenger-RNA splicing. Proc Natl Acad Sci USA 1991; 88: 10377–10381.
Knapp S, Yee LJ, Frodsham AJ, Hennig BJ, Hellier S, Zhang L et al. Polymorphisms in interferon-induced genes and the outcome of hepatitis C virus infection: roles of MxA, OAS-1 and PKR. Genes Immun 2003; 4: 411–419.
Wang SS, Gonzalez P, Yu K, Porras C, Li Q, Safaeian M et al. Common genetic variants and risk for HPV persistence and progression to cervical cancer. PLoS ONE 2010; 5: e8667.
He J, Feng D, de Vlas SJ, Wang H, Fontanet A, Zhang P et al. Association of SARS susceptibility with single nucleic acid polymorphisms of OAS1 and MxA genes: a case-control study. BMC Infect Dis 2006; 6: 106.
Tessier MC, Qu HQ, Frechette R, Bacot F, Grabs R, Taback SP et al. Type 1 diabetes and the OAS gene cluster: association with splicing polymorphism or haplotype? J Med Genet 2006; 43: 129–132.
Barkhash AV, Perelygin AA, Babenko VN, Myasnikova NG, Pilipenko PI, Romaschenko AG et al. Variability in the 2'-5'-oligoadenylate synthetase gene cluster is associated with human predisposition to tick-borne encephalitis virus-induced disease. J Infect Dis 2010; 202: 1813–1818.
Acknowledgements
This research was supported by the Global Research Laboratory Program of the National Research Foundation (NRF) funded by the Ministry of Science, ICT and Future Planning (MEST, K20705000006-12A0500-00610 to Y-JK), the Bio and Medical Technology Development Program of the National Research Foundation (NRF) funded by the Ministry of Science, ICT and Future Planning (MEST, 2012028272 to Y-JK) and the Korea Health Technology R&D Project through the Korea Health Industry Development Institute (KHIDI), funded by the Ministry of Health and Welfare, Republic of Korea (grant number: HI13C08470200).
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
This work is licensed under a Creative Commons Attribution 3.0 Unported License. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in the credit line; if the material is not included under the Creative Commons license, users will need to obtain permission from the license holder to reproduce the material. To view a copy of this license, visit http://creativecommons.org/licenses/by/3.0/
About this article
Cite this article
Choi, U., Kang, JS., Hwang, Y. et al. Oligoadenylate synthase-like (OASL) proteins: dual functions and associations with diseases. Exp Mol Med 47, e144 (2015). https://doi.org/10.1038/emm.2014.110
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/emm.2014.110
This article is cited by
-
Revealing the changes in signaling pathways caused by tofacitinib in patients with rheumatoid arthritis through RNA sequencing and the correlation with clinical parameters
Clinical Rheumatology (2024)
-
ROGUE: an R Shiny app for RNA sequencing analysis and biomarker discovery
BMC Bioinformatics (2023)
-
Occludin Regulates HIV-1 Infection by Modulation of the Interferon Stimulated OAS Gene Family
Molecular Neurobiology (2023)
-
Cytosolic nucleic acid sensing and mitochondrial transcriptomic changes as early triggers of metabolic disease in db/db mice
Mammalian Genome (2023)
-
Overexpression of OASL upregulates TET1 to induce aberrant activation of CD4+ T cells in systemic sclerosis via IRF1 signaling
Arthritis Research & Therapy (2022)