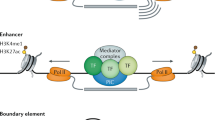
Abstract
Whole-genome sequence assemblies are now available for seven different animals, including nematode worms, mice and humans. Comparative genome analyses reveal a surprising constancy in genetic content: vertebrate genomes have only about twice the number of genes that invertebrate genomes have, and the increase is primarily due to the duplication of existing genes rather than the invention of new ones. How, then, has evolutionary diversity arisen? Emerging evidence suggests that organismal complexity arises from progressively more elaborate regulation of gene expression.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
References
Ruvkun, G. & Hobert, O. The taxonomy of developmental control in Caenorhabditis elegans. Science 282, 2033–2041 (1998)
Adams, M. D. et al. The genome sequence of Drosophila melanogaster. Science 287, 2185–2195 (2000)
Baltimore, D. Our genome unveiled. Nature 409, 814–816 (2001)
Lander, E. S. et al. Initial sequencing and analysis of the human genome. Nature 409, 860–921 (2001)
Graveley, B. R. Alternative splicing: increasing diversity in the proteomic world. Trends Genet. 17, 100–107 (2001)
Agrawal, A., Eastman, Q. M. & Schatz, D. G. Transposition mediated by RAG1 and RAG2 and its implications for the evolution of the immune system. Nature 394, 744–751 (1998)
Harafuji, N., Keys, D. N. & Levine, M. Genome-wide analysis of tissue-specific enhancers in the Ciona tadpole. Proc. Natl Acad. Sci. 99, 6802–6805 (2002)
Davidson, E. H. Genomic Regulatory Systems: Development and Evolution (Academic, New York, 2001)
Wyrick, J. J. & Young, R. A. Deciphering gene expression regulatory networks. Curr. Opin. Genet. Dev. 12, 130–136 (2002)
Aoyagi, N. & Wassarman, D. A. Genes encoding Drosophila melanogaster RNA polymerase II general transcription factors: diversity in TFIIA and TFIID components contributes to gene-specific transcription regulation. J. Cell Biol. 150, F45–F50 (2000)
Struhl, K., Kadosh, D., Keaveney, M., Kuras, L. & Moqtaderi, Z. Activation and repression mechanisms in yeast. Cold Spring Harb. Symp. Quant Biol 63, 413–421 (1998)
de Bruin, D., Zaman, Z., Liberatore, R. A. & Ptashne, M. Telomere looping permits gene activation by a downstream UAS in yeast. Nature 409, 109–113 (2001)
Brand, A. H., Breeden, L., Abraham, J., Sternglanz, R. & Nasmyth, K. Characterization of a “silencer” in yeast: a DNA sequence with properties opposite to those of a transcriptional enhancer. Cell 41, 41–48 (1985)
Smale, S. T. & Kadonaga, J. T. The RNA polymerase II core promoter. Annu. Rev. Biochem. (in the press)
Su, W., Jackson, S., Tjian, R. & Echols, H. DNA looping between sites for transcriptional activation: self-association of DNA-bound Sp1. Genes Dev. 5, 820–826 (1991)
Calhoun, V. C., Stathopoulos, A. & Levine, M. Promoter-proximal tethering elements regulate enhancer-promoter specificity in the Drosophila Antennapedia complex. Proc. Natl Acad. Sci. USA 99, 9243–9247 (2002)
Burgess-Beusse, B. et al. The insulation of genes from external enhancers and silencing chromatin. Proc. Natl Acad. Sci. USA 99, 16433–16437 (2002)
Banerji, J., Rusconi, S. & Schaffner, W. Expression of a beta-globin gene is enhanced by remote SV40 DNA sequences. Cell 27, 299–308 (1981)
DiMattia, G. E. et al. The Pit-1 gene is regulated by distinct early and late pituitary-specific enhancers. Dev. Biol. 182, 180–190 (1997)
Vesque, C. et al. Hoxb-2 transcriptional activation in rhombomeres 3 and 5 requires an evolutionarily conserved cis-acting element in addition to the Krox-20 binding site. EMBO J. 15, 5383–5396 (1996)
Genetta, T., Ruezinsky, D. & Kadesch, T. Displacement of an E-box-binding repressor by basic helix-loop-helix proteins: implications for B-cell specificity of the immunoglobulin heavy-chain enhancer. Mol. Cell Biol. 14, 6153–6163 (1994)
Webber, A. L., Ingram, R. S., Levorse, J. M. & Tilghman, S. M. Location of enhancers is essential for the imprinting of H19 and Igf2 genes. Nature 391, 711–715 (1998)
Leighton, P. A., Saam, J. R., Ingram, R. S., Stewart, C. L. & Tilghman, S. M. An enhancer deletion affects both H19 and Igf2 expression. Genes Dev. 9, 2079–2089 (1995)
Dorsett, D. Distant liaisons: long-range enhancer-promoter interactions in Drosophila. Curr. Opin. Genet. Dev. 9, 505–514 (1999)
Merli, C., Bergstrom, D. E., Cygan, J. A. & Blackman, R. K. Promoter specificity mediates the independent regulation of neighboring genes. Genes Dev. 10, 1260–1270 (1996)
DiLeone, R. J., Russell, L. B. & Kingsley, D. M. An extensive 3′ regulatory region controls expression of Bmp5 in specific anatomical structures of the mouse embryo. Genetics 148, 401–408 (1998)
Small, S., Arnosti, D. N. & Levine, M. Spacing ensures autonomous expression of different stripe enhancers in the even-skipped promoter. Development 119, 762–772 (1993)
Fujioka, M., Emi-Sarker, Y., Yusibova, G. L., Goto, T. & Jaynes, J. B. Analysis of an even-skipped rescue transgene reveals both composite and discrete neuronal and early blastoderm enhancers, and multi-stripe positioning by gap gene repressor gradients. Development 126, 2527–2538 (1999)
Mannervik, M., Nibu, Y., Zhang, H. & Levine, M. Transcriptional coregulators in development. Science 284, 606–609 (1999)
Schroder, C., Tautz, D., Seifert, E. & Jackle, H. Differential regulation of the two transcripts from the Drosophila gap segmentation gene hunchback. EMBO J. 7, 2881–2887 (1988)
Bell, A. C., West, A. G. & Felsenfeld, G. Insulators and boundaries: versatile regulatory elements in the eukaryotic genome. Science 291, 447–450 (2001)
Kellum, R. & Schedl, P. A position-effect assay for boundaries of higher order chromosomal domains. Cell 64, 941–950 (1991)
Choi, O. R. & Engel, J. D. Developmental regulation of beta-globin gene switching. Cell 55, 17–26 (1988)
Ohtsuki, S., Levine, M. & Cai, H. N. Different core promoters possess distinct regulatory activities in the Drosophila embryo. Genes Dev. 12, 547–556 (1998)
Butler, J. E. & Kadonaga, J. T. Enhancer-promoter specificity mediated by DPE or TATA core promoter motifs. Genes Dev. 15, 2515–2519 (2001)
Perkins, A. C., Gaensler, K. M. & Orkin, S. H. Silencing of human fetal globin expression is impaired in the absence of the adult beta-globin gene activator protein EKLF. Proc. Natl Acad. Sci. 93, 12267–12271 (1996)
Roeder, R. G. Role of general and gene-specific cofactors in the regulation of eukaryotic transcription. Cold Spring Harb. Symp Quant. Biol. 63, 201–218 (1998)
Freiman, R. N. et al. Requirement of tissue-selective TBP-associated factor TAFII105 in ovarian development. Science 293, 2084–2087 (2001)
Hiller, M. A., Lin, T. Y., Wood, C. & Fuller, M. T. Developmental regulation of transcription by a tissue-specific TAF homolog. Genes Dev. 15, 1021–1030 (2001)
Holmes, M. & Tjian, R. Promoter selective properties of the TBP-related factor TRF1. Science 288, 867–870 (2000)
Rabenstein, M. D., Zhou, S., Lis, J. T. & Tjian, R. TATA box-binding protein (TBP)-related factor 2 (TRF2), a third member of the TBP family. Proc. Natl Acad. Sci. USA 96, 4791–4796 (1999)
Kaltenbach, L., Horner, M. A., Rothman, J. H. & Mango, S. E. The TBP-like factor CeTLF is required to activate RNA polymerase II transcription during C. elegans embryogenesis. Mol. Cell 6, 705–713 (2000)
Veenstra, G. J., Weeks, D. L. & Wolffe, A. P. Distinct roles for TBP and TBP-like factor in early embryonic gene transcription in Xenopus. Science 290, 2312–2315 (2000)
Zhang, D., Penttila, T. L., Morris, P. L., Teichmann, M. & Roeder, R. G. Spermiogenesis deficiency in mice lacking the Trf2 gene. Science 292, 1153–1155 (2001)
Martianov, I. et al. Late arrest of spermiogenesis and germ cell apoptosis in mice lacking the TBP-like TLF/TRF2 gene. Mol. Cell 7, 509–515 (2001)
Hochheimer, A., Zhou, S., Zheng, S., Holmes, M. & Tjian, R. Promoter Selectivity and target gene identification of a DREF-containing TRF2 complex. Nature 420, 439–445 (2002)
Kim, Y.-J., Bjorklund, S., Li, Y., Sayre, M. H. & Kornberg, R. D. A multiprotein mediator of transcriptional activation and its interaction with the C-terminal repeat domain of RNA polymerase II. Cell 77, 599–608 (1994)
Koleske, A. J. & Young, R. A. An RNA polymerase II holoenzyme responsive to activators. Nature 368, 466–469 (1994)
Fondell, J. D., Ge, H. & Roeder, R. G. Ligand induction of a transcriptionally active thyroid hormone receptor coactivator complex. Proc. Natl Acad. Sci. USA 93, 8329–8333 (1996)
Rachez, C. et al. Ligand-dependent transcription activation by nuclear receptors requires the DRIP complex. Nature 398, 824–828 (1999)
Gu, W. et al. A novel human SRB/MED-containing cofactor complex, SMCC, involved in transcription regulation. Mol. Cell 3, 97–108 (1999)
Akoulitchev, S., Chuikov, S. & Reinberg, D. TFIIH is negatively regulated by cdk8-containing mediator complexes. Nature 407, 102–106 (2000)
Taatjes, D. J., Naar, A. M., Andel, F., Nogales, E. & Tjian, R. Structure, function, and activator-induced conformations of the CRSP co-activators. Science 295, 1058–1062 (2002)
Naar, A. M. et al. Composite co-activator ARC mediates chromatin-directed transcriptional activation. Nature 398, 828–832 (1999)
Ryu, S., Zhou, S., Ladurner, A. G. & Tjian, R. The transcriptional cofactor complex CRSP is required for activity of the enhancer-binding protein Sp1. Nature 397, 446–450 (1999)
Ito, M. & Roeder, R. G. The TRAP/SMCC/Mediator complex and thyroid hormone receptor function. Trends Endocrinol. Metab. 12, 127–134 (2001)
Dotson, M. R. et al. Structural organization of yeast and mammalian mediator complexes. Proc. Natl Acad. Sci. USA 97, 14307–14310 (2000)
Malik, S. & Roeder, R. G. Transcriptional regulation through Mediator-like coactivators in yeast and metazoan cells. Trends Biochem. Sci. 25, 277–283 (2000)
Glass, C. K. & Rosenfeld, M. G. The coregulator exchange in transcriptional functions of nuclear receptors. Genes Dev. 14, 121–141 (2000)
Bromberg, J. & Chen, X. STAT proteins: signal transducers and activators of transcription. Methods Enzymol. 333, 138–151 (2001)
Wang, W. et al. Diversity and specialization of mammalian SWI/SNF complexes. Genes Dev. 10, 2117–2130 (1996)
Goodman, R. H. & Smolik, S. CBP/p300 in cell growth, transformation, and development. Genes Dev. 14, 1553–1577 (2000)
Tamkun, J. W. et al. Brahma: a regulator of Drosophila homeotic genes structurally related to the yeast transcriptional activator SNF2/SWI2. Cell 68, 561–572 (1992)
Khavari, P. A., Peterson, C. L., Tamkun, J. W., Mendel, D. B. & Crabtree, G. R. BRG1 contains a conserved domain of the SWI2/SNF2 family necessary for normal mitotic growth and transcription. Nature 366, 170–174 (1993)
Feng, Q. & Zhang, Y. The NuRD complex: linking histone modification to nucleosome remodeling. Curr. Top. Microbiol. Immunol. 274, 269–290 (2003)
Tsukiyama, T., Daniel, C., Tamkun, J. & Wu, C. ISWI, a member of the SWI2/SNF2 ATPase family, encodes the 140 kDa subunit of thenucleosome remodeling factor. Cell 83, 1021–1026 (1995)
LeRoy, G., Loyola, A., Lane, W. S. & Reinberg, D. Purification and characterization of a human factor that assembles and remodels chromatin. J. Biol. Chem. 275, 14787–14790 (2000)
Olave, I. A., Reck-Peterson, S. L. & Crabtree, G. R. Nuclear actin and actin-related proteins in chromatin remodeling. Annu. Rev. Biochem. 71, 755–781 (2002)
Kehle, J. et al. dMi-2, a hunchback-interacting protein that functions in polycomb repression. Science 282, 1897–1900 (1998)
Markstein, M., Markstein, P., Markstein, V. & Levine, M. Genome-wide analysis of clustered Dorsal binding sites identifies putative target genes in the Drosophila embryo. Proc. Natl Acad. Sci. USA 99, 763–768 (2002)
Berman, B. P. et al. Exploiting transcription factor binding site clustering to identify cis-regulatory modules involved in pattern formation in the Drosophila genome. Proc. Natl Acad. Sci. USA 99, 757–762 (2002)
Rebeiz, M., Reeves, N. L. & Posakony, J. W. SCORE: A computational approach to the identification of cis-regulatory modules and target genes in whole-genome sequence data. Proc. Natl Acad. Sci. USA (in the press)
Halfon, M. S., Grad, Y., Church, G. M. & Michelson, A. M. Computation-based discovery of related transcriptional regulatory modules and motifs using an experimentally validated combinatorial model. Genome Res. 12, 1019–1028 (2002)
Rajewsky, N., Vergassola, M., Gaul, U. & Siggia, E. D. Computational detection of genomic cis-regulatory modules applied to body patterning in the early Drosophila embryo. BMC Bioinform. 3, 30 (2002)
Acknowledgements
We thank Y. Nibu, A. Ladurner and J. Ziegalbauer for preparing the figures. We also thank D. Rio and L. Mirels for critically reviewing the manuscript. M.L is supported by the NIH and R.T. is funded in part by a grant from the NIH.
Author information
Authors and Affiliations
Corresponding authors
Rights and permissions
About this article
Cite this article
Levine, M., Tjian, R. Transcription regulation and animal diversity. Nature 424, 147–151 (2003). https://doi.org/10.1038/nature01763
Issue Date:
DOI: https://doi.org/10.1038/nature01763
This article is cited by
-
Comparative phylogenetic analysis of the mediator complex subunit in asparagus bean (Vigna unguiculata ssp. sesquipedialis) and its expression profile under cold stress
BMC Genomics (2024)
-
Genome-wide identification and analysis of the growth-regulating factor (GRF) family in sweet cherry
Genetic Resources and Crop Evolution (2024)
-
Estimating transcriptome complexities across eukaryotes
BMC Genomics (2023)
-
Prediction of mammalian tissue-specific CLOCK–BMAL1 binding to E-box DNA motifs
Scientific Reports (2023)
-
Heat shock protein family A member 8 serving as a co-activator of transcriptional factor ETV4 up-regulates PHLDA2 to promote the growth of liver cancer
Acta Pharmacologica Sinica (2023)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.