Key Points
-
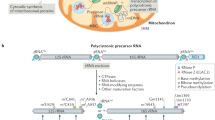
Proteomic studies revealed that mitochondria contain ∼1,000 (in yeast) to 1,500 (in humans) different proteins and led to the recent identification of new mitochondrial import pathways and components.
-
99% of mitochondrial proteins are encoded by nuclear genes, synthesized on cytosolic ribosomes and imported into mitochondria. 1% of mitochondrial proteins are encoded by the mitochondrial genome and synthesized in the mitochondrial matrix.
-
The translocase of the outer membrane (TOM) complex is the main entry gate used by most nucleus-encoded mitochondrial precursor proteins. Presequence-carrying preproteins are then imported by the presequence translocase of the inner membrane (TIM23) complex and the presequences are proteolytically removed by specific processing enzymes.
-
The precursors of hydrophobic metabolite carriers of the inner membrane are imported by the TOM complex, bind to chaperone-like proteins in the intermembrane space (small TIM proteins) and are inserted by the carrier translocase of the inner membrane (TIM22) complex.
-
Many proteins of the mitochondrial intermembrane space are small and contain characteristic Cys motifs. Most of these proteins are imported and folded by the redox-dependent mitochondrial intermembrane space assembly (MIA) machinery.
-
The mitochondrial outer membrane contains α-helical proteins and β-barrel proteins. Whereas the import pathways of α-helical proteins are only partly understood, the pathway for β-barrel proteins has been characterized and shown to require the TOM complex, small TIM chaperones of the intermembrane space and the sorting and assembly machinery (SAM) complex of the outer membrane.
Abstract
Mitochondria contain ∼1,000 different proteins, most of which are imported from the cytosol. Two import pathways that direct proteins into the mitochondrial inner membrane and matrix have been known for many years. The identification of numerous new transport components in recent proteomic studies has led to novel mechanistic insight into these pathways and the discovery of new import pathways into the outer membrane and intermembrane space. Protein translocases do not function as independent units but are integrated into dynamic networks and are connected to machineries that function in bioenergetics, mitochondrial morphology and coupling to the endoplasmic reticulum.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$189.00 per year
only $15.75 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Dolezal, P., Likic, V., Tachezy, J. & Lithgow, T. Evolution of the molecular machines for protein import into mitochondria. Science 313, 314–318 (2006).
Sickmann, A. et al. The proteome of Saccharomyces cerevisiae mitochondria. Proc. Natl Acad. Sci. USA 100, 13207–13212 (2003).
Pagliarini, D. J. et al. A mitochondrial protein compendium elucidates complex I disease biology. Cell 134, 112–123 (2008).
Lill, R. Function and biogenesis of iron-sulphur proteins. Nature 460, 831–838 (2009).
Neupert, W. & Herrmann, J. M. Translocation of proteins into mitochondria. Annu. Rev. Biochem. 76, 723–749 (2007). A detailed overview of the protein sorting pathways of mitochondria.
Saitoh, T. et al. Tom20 recognizes mitochondrial presequences through dynamic equilibrium among multiple bound states. EMBO J. 26, 4777–4787 (2007).
Chacinska, A., Koehler, C. M., Milenkovic, D., Lithgow, T. & Pfanner, N. Importing mitochondrial proteins: machineries and mechanisms. Cell 138, 628–644 (2009).
Walther, D. M. & Rapaport, D. Biogenesis of mitochondrial outer membrane proteins. Biochim. Biophys. Acta 1793, 42–51 (2009).
Rehling, P. et al. Protein insertion into the mitochondrial inner membrane by a twin-pore translocase. Science 299, 1747–1751 (2003).
Webb, C. T., Gorman, M. A., Lazarou, M., Ryan, M. T. & Gulbis, J. M. Crystal structure of the mitochondrial chaperone TIM9•10 reveals a six-bladed α-propeller. Mol. Cell 21, 123–133 (2006). Describes a high resolution structure of the small TIM chaperone complex of the mitochondrial intermembrane space that probably binds hydrophobic precursors of outer and inner membrane proteins by tentacle-like protrusions.
Koehler, C. M. New developments in mitochondrial assembly. Annu. Rev. Cell. Dev. Biol. 20, 309–335 (2004).
Chacinska, A. et al. Essential role of Mia40 in import and assembly of mitochondrial intermembrane space proteins. EMBO J. 23, 3735–3746 (2004). Identification of the MIA machinery.
Naoé, M. et al. Identification of Tim40 that mediates protein sorting to the mitochondrial intermembrane space. J. Biol. Chem. 279, 47815–47821 (2004).
Terziyska. et al. Mia40, a novel factor for protein import into the intermembrane space of mitochondria is able to bind metal ions. FEBS Lett. 579, 179–184 (2005).
Wiedemann, N. et al. Machinery for protein sorting and assembly in the mitochondrial outer membrane. Nature 424, 565–571 (2003).
Kozjak, V. et al. An essential role of Sam50 in the protein sorting and assembly machinery of the mitochondrial outer membrane. J. Biol. Chem. 278, 48520–48523 (2003).
Paschen, S. A. et al. Evolutionary conservation of biogenesis of β-barrel membrane proteins. Nature 426, 862–866 (2003).
Gentle, I., Gabriel, K., Beech, P., Waller, R. & Lithgow, T. The Omp85 family of proteins is essential for outer membrane biogenesis in mitochondria and bacteria. J. Cell Biol. 164, 19–24 (2004).
Geissler, A. et al. The mitochondrial presequence translocase: an essential role of Tim50 in directing preproteins to the import channel. Cell 111, 507–518 (2002).
Frazier, A. E. et al. Pam16 has an essential role in the mitochondrial protein import motor. Nature Struct. Mol. Biol. 11, 226–233 (2004).
Kozany, C., Mokranjac, D., Sichting, M., Neupert, W. & Hell, K. The J domain-related cochaperone Tim16 is a constituent of the mitochondrial TIM23 preprotein translocase. Nature Struct. Mol. Biol. 11, 234–241 (2004).
Wittig, I., Braun, H. P. & Schägger, H. Blue native PAGE. Nature Protoc. 1, 418–428 (2006).
Chacinska, A. et al. Mitochondrial presequence translocase: switching between TOM tethering and motor recruitment involves Tim21 and Tim17. Cell 120, 817–829 (2005).
Wiedemann, N., van der Laan, M., Hutu, D. P., Rehling, P. & Pfanner, N. Sorting switch of mitochondrial presequence translocase involves coupling of motor module to respiratory chain. J. Cell Biol. 179, 1115–1122 (2007).
Dienhart, M. K. & Stuart, R. A. The yeast Aac2 protein exists in physical association with the cytochrome bc1–COX supercomplex and the TIM23 machinery. Mol. Biol. Cell 19, 3934–3943 (2008).
Reinders, J., Zahedi, R. P., Pfanner, N., Meisinger, C. & Sickmann, A. Toward the complete yeast mitochondrial proteome: multidimensional separation techniques for mitochondrial proteomics. J. Proteome Res. 5, 1543–1554 (2006). Together with references 2 and 3, this paper reports on a comprehensive analysis of the mitochondrial proteome, revealing a large range of mitochondrial functions.
Meisinger, C., Sickmann, A. & Pfanner, N. The mitochondrial proteome: from inventory to function. Cell 134, 22–24 (2009).
Vögtle, F. N. et al. Global analysis of the mitochondrial N-proteome identifies a processing peptidase critical for protein stability. Cell 139, 428–439 (2009). Systematic analysis of mitochondrial presequences and their processing identifies an intermediate cleaving peptidase that regulates mitochondrial protein stability.
Li, J., Qian, X., Hu, J. & Sha, B. Molecular chaperone Hsp70/Hsp90 prepares the mitochondrial outer membrane translocon receptor Tom71 for preprotein loading. J. Biol. Chem. 284, 23852–23859 (2009).
Yamamoto. et al. Tim50 is a subunit of the TIM23 complex that links protein translocation across the outer and inner mitochondrial membranes. Cell 111, 519–528 (2002).
Tamura, Y. et al. Tim23–Tim50 pair coordinates functions of translocators and motor proteins in mitochondrial protein import. J. Cell Biol. 184, 129–141 (2009).
Mokranjac, D. et al. Role of Tim50 in the transfer of precursor proteins from the outer to inner membrane of mitochondria. Mol. Biol. Cell. 20, 1400–1407 (2009).
Meinecke, M. et al. Tim50 maintains the permeability barrier of the mitochondrial inner membrane. Science 312, 1523–1526 (2006).
Truscott, K. N. et al. A presequence- and voltage-sensitive channel of the mitochondrial preprotein translocase formed by Tim23. Nature Struct. Biol. 8, 1074–1082 (2001).
Gevorkyan-Airapetov, L. et al. Interaction of Tim23 with Tim50 is essential for protein translocation by the mitochondrial TIM23 complex. J. Biol. Chem. 284, 4865–4872 (2009).
Mokranjac, D., Popov-Celeketic, D., Hell, K. & Neupert, W. Role of Tim21 in mitochondrial translocation contact sites. J. Biol. Chem. 280, 23437–23440 (2005).
Albrecht, R. et al. The Tim21 binding domain connects the preprotein translocases of both mitochondrial membranes. EMBO Rep. 7, 1233–1238 (2006).
van der Laan, M. et al. Motor-free mitochondrial presequence translocase drives membrane integration of preproteins. Nature Cell Biol. 9, 1152–1159 (2007).
Chacinska, A. et al. Distinct forms of mitochondrial TOM–TIM supercomplexes define signal-dependent states of preprotein sorting. Mol. Cell. Biol. 30, 307–318 (2010).
van der Laan, M. et al. A role for Tim21 in membrane-potential-dependent preprotein sorting in mitochondria. Curr. Biol. 16, 2271–2276 (2006).
Li, Y. et al. The presequence translocase-associated protein import motor of mitochondria: Pam16 functions in an antagonistic manner to Pam18. J. Biol. Chem. 279, 38047–38054 (2004).
Mokranjac, D., Bourenkov, G., Hell, K., Neupert, W. & Groll, M. Structure and function of Tim14 and Tim16, the J and J-like components of the mitochondrial protein import motor. EMBO J. 25, 4675–4685 (2006).
D'Silva, P. R., Schilke, B., Hayashi, M. & Craig, E. A. Interaction of the J-protein heterodimer Pam18/Pam16 of the mitochondrial import motor with the translocon of the inner membrane. Mol. Biol. Cell 19, 424–432 (2008).
van der Laan, M. et al. Pam17 is required for architecture and translocation activity of the mitochondrial protein import motor. Mol. Cell. Biol. 25, 7449–7458 (2005).
Popov-Celeketic, D., Mapa, K., Neupert, W. & Mokranjac, D. Active remodeling of the TIM23 complex during translocation of preproteins into mitochondria. EMBO J. 27, 1469–1480 (2008).
Taylor, A. B. et al. Crystal structures of mitochondrial processing peptidase reveal the mode for specific cleavage of import signal sequences. Structure 9, 615–625 (2001). Describes a high resolution structure of MPP with presequence peptides in an extended conformation, in contrast to the α-helical conformation of presequence peptides bound to the receptor Tom20 described in reference 6, and shows that the conformation of presequences changes during import.
Gakh, O., Cavadini, P. & Isaya, G. Mitochondrial processing peptidases. Biochim. Biophys. Acta 1592, 63–77 (2002).
Esser, K., Tursun, B., Ingenhoven, M., Michaelis, G. & Pratje, E. A novel two-step mechanism for removal of a mitochondrial signal sequence involves the mAAA complex and the putative rhomboid protease Pcp1. J. Mol. Biol. 323, 835–843 (2002). Identification of a new pathway for processing mitochondrial presequences, involving an ATP-dependent protease of the inner membrane and the rhomboid protease Pcp1.
Luo, W., Fang, H. & Green, N. Substrate specificity of inner membrane peptidase in yeast mitochondria. Mol. Genet. Genomics 275, 431–436 (2006).
Koppen, M. & Langer, T. Protein degradation within mitochondria: versatile activities of AAA proteases and other peptidases. Crit. Rev. Biochem. Mol. Biol. 42, 221–242 (2007).
Herlan, M., Bornhövd, C., Hell, K., Neupert, W. & Reichert, A. Alternative topogenesis of Mgm1 and mitochondrial morphology depend on ATP and a functional import motor. J. Cell Biol. 165, 167–173 (2004).
Naamati, A., Regev-Rudzki, N., Galperin, S., Lill, R. & Pines, O. Dual targeting of Nfs1 and discovery of its novel processing enzyme, Icp55. J. Biol. Chem. 284, 30200–30208 (2009).
Mogk, A., Schmidt, R. & Bukau, B. The N-end rule pathway for regulated proteolysis: prokaryotic and eukaryotic strategies. Trends Cell. Biol. 17, 165–172 (2007).
Varshavsky, A. The N-end rule at atomic resolution. Nature Struct. Mol. Biol. 15, 1238–1240 (2008).
Young, J. C., Hoogenraad, N. J. & Hartl, F. U. Molecular chaperones Hsp90 and Hsp70 deliver preproteins to the mitochondrial import receptor Tom70. Cell 112, 41–50 (2003). Shows that the cytosolic chaperones Hsp70 and Hsp90 deliver precursor proteins to mitochondria and directly interact with the membrane-bound receptor Tom70.
Zara, V., Ferramosca, A., Robitaille-Foucher, P., Palmieri, F. & Young, J. C. Mitochondrial carrier protein biogenesis: role of the chaperones Hsc70 and Hsp90. Biochem. J. 419, 369–375 (2009).
Wiedemann, N., Pfanner, N. & Ryan, M. T. The three modules of ADP/ATP carrier cooperate in receptor recruitment and translocation into mitochondria. EMBO J. 20, 951–960 (2001).
Curran, S. P., Leuenberger, D., Oppliger, W. & Koehler, C. M. The Tim9p–Tim10p complex binds to the transmembrane domains of the ADP/ATP carrier. EMBO J. 21, 942–953 (2002).
Davis, A. J., Alder, N. N., Jensen, R. E. & Johnson, A. E. The Tim9p/10p and Tim8p/13p complexes bind to specific sites on TIM23 during mitochondrial protein import. Mol. Biol. Cell 18, 475–486 (2007). References 58 and 59 describe the interaction of membrane protein precursors with the small TIM chaperones of the mitochondrial intermembrane space.
Roesch, K., Curran, S. P., Tranebjaerg, L. & Koehler, C. M. Human deafness dystonia syndrome is caused by a defect in assembly of the DDP1/TIMM8a-TIMM13 complex. Hum. Mol. Gen. 11, 477–486 (2002). Molecular characterization of the first human disease caused by a defect in the mitochondrial protein import machinery.
Wagner, K. et al. The assembly pathway of the mitochondrial carrier translocase involves four preprotein translocases. Mol. Cell. Biol. 28, 4251–4260 (2008).
Lithgow, T. & Schneider, A. Evolution of macromolecular import pathways in mitochondria, hydrogenosomes and mitosomes. Philos. Trans. R. Soc. Lond. B Biol. Sci. 365, 799–817 (2010).
Gabriel, K. et al. Novel mitochondrial intermembrane space proteins as substrates of the MIA import pathway. J. Mol. Biol. 365, 612–620 (2007).
Longen, S. et al. Systematic analysis of the twin Cx9C protein family. J. Mol. Biol. 393, 356–368 (2009).
Milenkovic, D. et al. Identification of the signal directing Tim9 and Tim10 into the intermembrane space of mitochondria. Mol. Biol. Cell 20, 2530–2539 (2009).
Sideris, D. P. et al. A novel intermembrane space-targeting signal docks cysteines onto Mia40 during mitochondrial oxidative folding. J. Cell Biol. 187, 1007–1022 (2009).
Kawano, S. et al. Structural basis of yeast Tim40/Mia40 as an oxidative translocator in the mitochondrial intermembrane space. Proc. Natl Acad. Sci. USA 106, 14403–14407 (2009).
Banci, L. et al. MIA40 is an oxidoreductase that catalyzes oxidative protein folding in mitochondria. Nature Struct. Mol. Biol. 16, 198–206 (2009). References 67 and 68 report the high resolution structure of Mia40, the core of the MIA machinery, and provide implications for the mechanism of oxidative protein folding.
Mesecke, N. et al. A disulfide relay system in the intermembrane space of mitochondria that mediates protein import. Cell 121, 1059–1069 (2005). Together with reference 12, this study characterizes the MIA machinery that functions as a disulphide relay.
Rissler, M. et al. The essential mitochondrial protein Erv1 cooperates with Mia40 in biogenesis of intermembrane space proteins. J. Mol. Biol. 353, 485–492 (2005).
Allen, S., Balabanidou, V., Sideris, D. P., Lisowsky, T. & Tokatlidis, K. Erv1 mediates the Mia40-dependent protein import pathway and provides a functional link to the respiratory chain by shuttling electrons to cytochrome c. J. Mol. Biol. 353, 937–944 (2005).
Bien, M. et al. Mitochondrial disulfide bond formation is driven by intersubunit electron transfer in Erv1 and proofread by glutathione. Mol. Cell 37, 516–528 (2010).
Dabir, D. V. et al. A role for cytochrome c and cytochrome c peroxidase in electron shuttling from Erv1. EMBO J. 26, 4801–4811 (2007).
Bihlmaier, K. et al. The disulfide relay system of mitochondria is connected to the respiratory chain. J. Cell Biol. 179, 389–395 (2007).
Stojanovski, D. et al. Mitochondrial protein import: precursor oxidation in a ternary complex with disulfide carrier and sulfhydryl oxidase. J. Cell Biol. 183, 195–202 (2008).
Walther, D. M., Papic, D., Bos, M. P., Tommassen, J. & Rapaport, D. Signals in bacterial β-barrel proteins are functional in eukaryotic cells for targeting to and assembly in mitochondria. Proc. Natl Acad. Sci. USA 106, 2531–2536 (2009). Bacterial β-barrel proteins can be assembled by the mitochondrial SAM complex, revealing conservation of the pathway from bacteria to mitochondria.
Wiedemann, N. et al. Biogenesis of the protein import channel Tom40 of the mitochondrial outer membrane: intermembrane space components are involved in an early stage of the assembly pathway. J. Biol. Chem. 279, 18188–18194 (2004).
Hoppins, S. C. & Nargang, F. E. The Tim8-Tim13 complex of Neurospora crassa functions in the assembly of proteins into both mitochondrial membranes. J. Biol. Chem. 279, 12396–12405 (2004).
Kutik, S. et al. Dissecting membrane insertion of mitochondrial β-barrel proteins. Cell 132, 1011–1024 (2008). Identification of the mitochondrial β-signal.
Chan, N. C. & Lithgow, T. The peripheral membrane subunits of the SAM complex function codependently in mitochondrial outer membrane biogenesis. Mol. Biol. Cell 19, 126–136 (2008). References 79 and 80 dissect precursor binding to and release from the SAM complex of the outer membrane.
Meisinger, C. et al. The mitochondrial morphology protein Mdm10 functions in assembly of the preprotein translocase of the outer membrane. Dev. Cell 7, 61–71 (2004).
Thornton, N. et al. Two modular forms of the mitochondrial sorting and assembly machinery are involved in biogenesis of α-helical outer membrane proteins. J. Mol. Biol. 396, 540–549 (2010).
Yamano, K., Tanaka-Yamano, S. & Endo, T. Mdm10 as a dynamic constituent of the TOB/SAM complex directs coordinated assembly of Tom40. EMBO Rep. 11, 187–193 (2010).
Wideman, J. G. et al. Roles of the Mdm10, Tom7, Mdm12, and Mmm1 proteins in the assembly of mitochondrial outer membrane proteins in Neurospora crassa. Mol. Biol. Cell 21, 1725–1736 (2010).
Popov-Celeketic, J., Waizenegger, T. & Rapaport, D. Mim1 functions in an oligomeric form to facilitate the integration of Tom20 into the mitochondrial outer membrane. J. Mol. Biol. 376, 671–680 (2008).
Becker, T. et al. Biogenesis of the mitochondrial TOM complex: Mim1 promotes insertion and assembly of signal-anchored receptors. J. Biol. Chem. 283, 120–127 (2008).
Hulett, J. M. et al. The transmembrane segment of Tom20 is recognized by Mim1 for docking to the mitochondrial TOM complex. J. Mol. Biol. 376, 694–704 (2008).
Mnaimneh, S. et al. Exploration of essential gene functions via titratable promoter alleles. Cell 118, 31–44 (2004).
Stojanovski, D., Guiard, B., Kozjak-Pavlovic, V., Pfanner, N. & Meisinger, C. Alternative function for the mitochondrial SAM complex in biogenesis of α-helical TOM proteins. J. Cell Biol. 179, 881–893 (2007).
Otera, H. et al. A novel insertion pathway of mitochondrial outer membrane proteins with multiple transmembrane segments. J. Cell Biol. 179, 1355–1363 (2007). Identification of mechanisms for the insertion of multi-spanning α-helical proteins into the mitochondrial outer membrane.
Setoguchi, K., Otera, H. & Mihara, K. Cytosolic factor- and TOM-independent import of C-tail-anchored mitochondrial outer membrane proteins. EMBO J. 25, 5635–5647 (2006).
Kemper, C. et al. Integration of tail-anchored proteins into the mitochondrial outer membrane does not require any known import components. J. Cell Sci. 121, 1990–1998 (2008).
Sogo, L. F. & Yaffe, M. P. Regulation of mitochondrial morphology and inheritance by Mdm10p, a protein of the mitochondrial outer membrane. J. Cell Biol. 126, 1361–1373 (1994).
Meisinger, C. et al. The morphology proteins Mdm12/Mmm1 function in the major β-barrel assembly pathway of mitochondria. EMBO J. 26, 2229–2239 (2007).
Boldogh, I. R. et al. A protein complex containing Mdm10p, Mdm12p, and Mmm1p links mitochondrial membranes and DNA to the cytoskeleton-based segregation machinery. Mol. Biol. Cell 14, 4618–4627 (2003).
Meisinger, C. et al. Mitochondrial protein sorting: differentiation of β-barrel assembly by Tom7-mediated segregation of Mdm10. J. Biol. Chem. 281, 22819–22826 (2006).
Garcia-Rodriguez, L. J. et al. Mitochondrial inheritance is required for MEN-regulated cytokinesis in budding yeast. Curr. Biol. 19, 1730–1735 (2009).
Youngman, M. J., Hobbs, A. E., Burgess, S. M., Srinivasan, M. & Jensen, R. E. Mmm2p, a mitochondrial outer membrane protein required for yeast mitochondrial shape and maintenance of mtDNA nucleoids. J. Cell Biol. 164, 677–688 (2004).
Kornmann, B. et al. An ER-mitochondrial tethering complex revealed by a synthetic biology screen. Science 325, 477–481 (2009).
Vance, J. E. Phospholipid synthesis in a membrane fraction associated with mitochondria. J. Biol. Chem., 7248–7256 (1990).
de Brito, O. M. & Scorrano, L. Mitofusin 2 tethers endoplasmic reticulum to mitochondria. Nature 456, 605–610 (2008). References 99–101 identify protein-mediated interactions between mitochondria and the ER, with implications for the transfer of lipids and Ca2+.
Wiedemann, N., Meisinger, C. & Pfanner, N. Cell biology: connecting organelles. Science 325, 403–404 (2009).
Jiang, N., Levavasseur, F., McCright, B., Shoubridge, E. A. & Hekimi, S. Mouse CLK-1 is imported into mitochondria by an unusual process that requires a leader sequence but no membrane potential. J. Biol. Chem. 276, 29218–29225 (2001).
Wagner, K. et al. Mitochondrial F1Fo-ATP synthase: the small subunits e and g associate with monomeric complexes to trigger dimerization. J. Mol. Biol. 392, 855–861 (2009).
Yogev, O., Karniely, S. & Pines, O. Translation-coupled translocation of yeast fumarase into mitochondria in vivo. J. Biol. Chem. 282, 29222–29229 (2007).
Kellems, R. E., Allison, V. F. & Butow, R. A. Cytoplasmic type 80S ribosomes associated with yeast mitochondria IV: attachment of ribosomes to the outer membrane of isolated mitochondria. J. Cell Biol. 65, 14–81 (1975).
Marc, P. et al. Genome-wide analysis of mRNAs targeted to yeast mitochondria. EMBO Rep. 3, 159–164 (2002).
Eliyahu, E. et al. Tom20 mediates localization of mRNAs to mitochondria in a translation-dependent manner. Mol. Cell Biol. 30, 284–294 (2010).
Zahedi, R. P. et al. Proteomic analysis of the yeast mitochondrial outer membrane reveals accumulation of a subclass of preproteins. Mol. Biol. Cell 17, 1436–1450 (2006).
Gallas, M. R., Dienhart, M. K., Stuart, R. A. & Long, R. M. Characterization of Mmp37p, a Saccharomyces cerevisiae mitochondrial matrix protein with a role in mitochondrial protein import. Mol. Biol. Cell 17, 4051–4062 (2006).
Tamura, Y. et al. Identification of Tam41 maintaining integrity of the TIM23 protein translocator complex in mitochondria. J. Cell Biol. 174, 631–637 (2006).
Kutik, S. et al. The translocator maintenance protein Tam41 is required for mitochondrial cardiolipin biosynthesis. J. Cell Biol. 183, 1213–1221 (2008).
Gebert, N. et al. Mitochondrial cardiolipin involved in outer-membrane protein biogenesis: implications for Barth syndrome. Curr. Biol. 19, 2133–2139 (2009).
Ahting, U. et al. The TOM core complex: the general protein import pore of the outer membrane of mitochondria. J. Cell Biol. 147, 959–968 (1999).
Model, K., Meisinger, C. & Kühlbrandt, W. Cryo-electron microscopy structure of a yeast mitochondrial preprotein translocase. J. Mol. Biol. 383, 1049–1057 (2008).
Baker, M. J., Frazier, A. E., Gulbis, J. M. & Ryan, M. T. Mitochondrial protein-import machinery: correlating structure with function. Trends Cell Biol. 17, 456–464 (2007).
Wallace, D. C. & Fan, W. The pathophysiology of mitochondrial disease as modeled in the mouse. Genes Dev. 23, 1714–1736 (2009).
Reinders, J. et al. Profiling phosphoproteins of yeast mitochondria reveals a role of phosphorylation in assembly of the ATP synthase. Mol. Cell. Proteomics 6, 1896–1906 (2007).
Lee, J. et al. Mitochondrial phosphoproteome revealed by an improved IMAC method and MS/MS/MS. Mol. Cell. Proteomics 6, 669–676 (2007).
Mootha, V. K. et al. Integrated analysis of protein composition, tissue diversity and gene regulation in mouse mitochondria. Cell 115, 629–640 (2003).
Schmitt, S. et al. Proteome analysis of mitochondrial outer membrane from Neurospora crassa. Proteomics 6, 72–80 (2006).
Timmer, J. C. et al. Profiling constitutive proteolytic events in vivo. Biochem. J. 407, 41–48 (2007).
Kruft, V., Eubel, H., Jänsch, L., Werhahn, W. & Braun, H. P. Proteomic approach to identify novel mitochondrial proteins in Arabidopsis. Plant Physiol. 127, 1694–1710 (2001).
Pflieger, D. et al. Systematic identification of mitochondrial proteins by LC-MS/MS. Anal. Chem. 74, 2400–2406 (2002).
Taylor, S. W. et al. Characterization of the human heart mitochondrial proteome. Nature Biotechnol. 21, 281–286 (2003).
Heazlewood, J. L. et al. Experimental analysis of the Arabidopsis mitochondrial proteome highlights signaling and regulatory components, provides assessment of targeting prediction programs, and indicates plant-specific mitochondrial proteins. Plant Cell 16, 241–256 (2004).
Prokisch, H. et al. Integrative analysis of the mitochondrial proteome in yeast. PLoS Biol. 2, e160 (2004).
Forner, F., Foster, L. J., Campanaro, S., Valle, G. & Mann, M. Quantitative proteomic comparison of rat mitochondria from muscle, heart, and liver. Mol. Cell. Proteomics 5, 608–619 (2006).
Smith, D. G. et al. Exploring the mitochondrial proteome of the ciliate protozoon Tetrahymena thermophila: direct analysis by tandem mass spectrometry. J. Mol. Biol. 374, 837–863 (2007).
Nash, R. et al. Expanded protein information at SGD: new pages and proteome browser. Nucleic Acids Res. 35, D468–D471 (2007).
Nolden, M. et al. The m-AAA protease defective in hereditary spastic paraplegia controls ribosome assembly in mitochondria. Cell 123, 277–289 (2005). An ATP-dependent protease of the mitochondrial inner membrane controls ribosome assembly by processing a preprotein, implicating the mechanism of axonal degeneration in hereditary spastic paraplegia.
Osman, C., Wilmes, C., Tatsuta, T. & Langer, T. Prohibitins interact genetically with Atp23, a novel processing peptidase and chaperone for the F1Fo-ATP synthase. Mol. Biol. Cell 18, 627–635 (2007).
Zeng, X., Neupert, W. & Tzagoloff, A. The metalloprotease encoded by ATP23 has a dual function in processing and assembly of subunit 6 of mitochondrial ATPase. Mol. Biol. Cell 18, 617–626 (2007).
Esser, K., Jan, P. S., Pratje, E. & Michaelis, G. The mitochondrial IMP peptidase of yeast: functional analysis of domains and identification of Gut2 as a new natural substrate. Mol. Genet. Genomics 271, 616–626 (2004).
Acknowledgements
The authors' research is supported by the Deutsche Forschungsgemeinschaft, Excellence Initiative of the German Federal and State Governments (EXC 294 BIOSS; GSC-4 Spemann Graduate School), Trinationales Graduiertenkolleg GRK 1478, Bundesministerium für Bildung und Forschung, Sonderforschungsbereich 746, Gottfried Wilhelm Leibniz Program, Landesforschungspreis Baden-Württemberg and Fonds der Chemischen Industrie.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Related links
Glossary
- α-proteobacterium
-
A Gram-negative (outer membrane-containing) bacterium, such as Rickettsia spp., that is probably the closest living bacterial relative of mitochondria.
- 70 kDa heat shock protein
-
An ATP-dependent molecular chaperone that is essential in unstressed and stressed cells. The chaperones bind hydrophobic segments of unfolded proteins, preventing protein aggregation and promoting protein transport and folding.
- Deafness dystonia syndrome
-
An X chromosome-linked neurodegenerative disease that includes deafness, cortical blindness and dystonia. It was the first human disease caused by a defect in the mitochondrial protein import machinery (specifically, in Tim8 (known as TIMM8A in humans)). It is also called Mohr–Tranebjaerg syndrome.
- Porin
-
A pore-forming protein of the mitochondrial outer membrane that is permeable for many metabolites. It is the most abundant outer membrane protein and is also called voltage-dependent anion channel (VDAC).
- Mitofusin
-
A mitochondrial outer membrane protein required for fusion of mitochondria and maintenance of mitochondrial morphology.
- Cardiolipin
-
A large, dimeric phospholipid that is a characteristic of mitochondria and consists of two phosphatidyl moieties linked by glycerol.
- Liposome
-
An artificial lipid vesicle that is typically formed by a phospholipid bilayer (membrane).
Rights and permissions
About this article
Cite this article
Schmidt, O., Pfanner, N. & Meisinger, C. Mitochondrial protein import: from proteomics to functional mechanisms. Nat Rev Mol Cell Biol 11, 655–667 (2010). https://doi.org/10.1038/nrm2959
Issue Date:
DOI: https://doi.org/10.1038/nrm2959
This article is cited by
-
Tom70-regulated mitochondrial biogenesis via TFAM improves hypoxia-induced dysfunction of pulmonary vascular endothelial cells and alleviates hypoxic pulmonary hypertension
Respiratory Research (2023)
-
Mitochondrial heterogeneity in diseases
Signal Transduction and Targeted Therapy (2023)
-
Nanomedicine in cancer therapy
Signal Transduction and Targeted Therapy (2023)
-
Identification of a novel C6 protein encoded by tomato leaf curl China virus
Phytopathology Research (2022)
-
Mitochondria homeostasis: Biology and involvement in hepatic steatosis to NASH
Acta Pharmacologica Sinica (2022)