Key Points
-
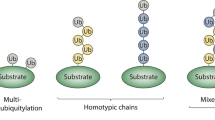
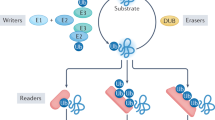
Ubiquitylation is mediated by the sequential activity of activating (E1), conjugating (E2) and ligating (E3) enzymes, resulting in the conjugation of monoubiquitin or polyubiquitin chains of different length and linkages to target proteins. Ubiquitin signals are decoded by ubiquitin receptors containing ubiquitin-binding domains (UBDs).
-
The ubiquitin network is strictly regulated in a spatiotemporal manner. Compartmentalization of ubiquitin-conjugation machinery proteins, deubiquitylating enzymes (DUBs) and downstream effectors, as well as sequential series of ubiquitylation events, are common strategies to confer spatial and temporal hierarchy to ubiquitylation.
-
Subcellular targeting of ubiquitin network components can be promoted by transmembrane anchors, nuclear localization signals, binding to scaffold proteins or reorganizations induced by other post-translational modifications. Coupled monoubiquitylation is also frequently used to control the activity and localization of proteins.
-
The nuclear factor-κB (NF-κB) pathway is regulated by many ubiquitin species, including Lys11-, Lys48- and Lys63-linked chains, as well as linear chains of different lengths. These function to spatially and temporally reorganize kinase complex activation in distinct branches of the pathway.
-
Ubiquitylation is a key determinant for spatial coordination of DNA repair and for the sorting and trafficking of membrane proteins by endosomal sorting complex required for transport (ESCRT)-associated proteins along the endocytic pathway. Ubiquitylation is also crucial for protein turnover in the ubiquitin–proteasome system, endoplasmic reticulum-associated degradation (ERAD) and autophagosomal pathways.
-
Spatial organization of ubiquitin-conjugating proteins and their regulators is crucial for pattern formation in the early development of many species, as well as during the specification and functional adaptation of individual cell and tissue types, in particular the nervous system.
Abstract
In the past decade, the diversity of signals generated by the ubiquitin system has emerged as a dominant regulator of biological processes and propagation of information in the eukaryotic cell. A wealth of information has been gained about the crucial role of spatial and temporal regulation of ubiquitin species of different lengths and linkages in the nuclear factor-κB (NF-κB) pathway, endocytic trafficking, protein degradation and DNA repair. This spatiotemporal regulation is achieved through sophisticated mechanisms of compartmentalization and sequential series of ubiquitylation events and signal decoding, which control diverse biological processes not only in the cell but also during the development of tissues and entire organisms.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$189.00 per year
only $15.75 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Weissman, A. M. Themes and variations on ubiquitylation. Nature Rev. Mol. Cell Biol. 2, 169–178 (2001).
Hershko, A. & Ciechanover, A. The ubiquitin system. Annu. Rev. Biochem. 67, 425–479 (1998).
Varshavsky, A. Regulated protein degradation. Trends Biochem. Sci. 30, 283–286 (2005).
Deshaies, R. J. & Joazeiro, C. A. RING domain E3 ubiquitin ligases. Annu. Rev. Biochem. 78, 399–434 (2009).
Ye, Y. & Rape, M. Building ubiquitin chains: E2 enzymes at work. Nature Rev. Mol. Cell Biol. 10, 755–764 (2009).
Schulman, B. A. & Harper, J. W. Ubiquitin-like protein activation by E1 enzymes: the apex for downstream signalling pathways. Nature Rev. Mol. Cell Biol. 10, 319–331 (2009).
Ikeda, F. & Dikic, I. Atypical ubiquitin chains: new molecular signals. 'Protein modifications: beyond the usual suspects' review series. EMBO Rep. 9, 536–542 (2008).
Iwai, K. & Tokunaga, F. Linear polyubiquitination: a new regulator of NF-κB activation. EMBO Rep. 10, 706–713 (2009).
Koegl, M. et al. A novel ubiquitination factor, E4, is involved in multiubiquitin chain assembly. Cell 96, 635–644 (1999).
Hoppe, T. Multiubiquitylation by E4 enzymes: 'one size' doesn't fit all. Trends Biochem. Sci. 30, 183–187 (2005).
Dikic, I., Wakatsuki, S. & Walters, K. J. Ubiquitin-binding domains — from structures to functions. Nature Rev. Mol. Cell Biol. 10, 659–671 (2009).
Wilkinson, K. D. DUBs at a glance. J. Cell Sci. 122, 2325–2329 (2009).
Komander, D., Clague, M. J. & Urbe, S. Breaking the chains: structure and function of the deubiquitinases. Nature Rev. Mol. Cell Biol. 10, 550–563 (2009).
Ravid, T. & Hochstrasser, M. Diversity of degradation signals in the ubiquitin-proteasome system. Nature Rev. Mol. Cell Biol. 9, 679–690 (2008).
Finley, D. Recognition and processing of ubiquitin-protein conjugates by the proteasome. Annu. Rev. Biochem. 78, 477–513 (2009).
Ulrich, H. D. & Walden, H. Ubiquitin signalling in DNA replication and repair. Nature Rev. Mol. Cell Biol. 11, 479–489 (2010).
Hirsch, C., Gauss, R., Horn, S. C., Neuber, O. & Sommer, T. The ubiquitylation machinery of the endoplasmic reticulum. Nature 458, 453–460 (2009).
Raiborg, C. & Stenmark, H. The ESCRT machinery in endosomal sorting of ubiquitylated membrane proteins. Nature 458, 445–452 (2009).
Wertz, I. E. & Dixit, V. M. Regulation of death receptor signaling by the ubiquitin system. Cell Death Differ. 17, 14–24 (2010).
Kirkin, V., McEwan, D. G., Novak, I. & Dikic, I. A role for ubiquitin in selective autophagy. Mol. Cell 34, 259–269 (2009).
Schwartz, A. L., Ciechanover, A., Brandt, R. A. & Geuze, H. J. Immunoelectron microscopic localization of ubiquitin in hepatoma cells. EMBO J. 7, 2961–2966 (1988).
Beers, E. P., Moreno, T. N. & Callis, J. Subcellular localization of ubiquitin and ubiquitinated proteins in Arabidopsis thaliana. J. Biol. Chem. 267, 15432–15439 (1992).
Ikeda, F., Crosetto, N. & Dikic, I. What determines the specificity and outcomes of ubiquitin signaling? Cell 143, 677–681 (2010).
Bodine, S. C. et al. Identification of ubiquitin ligases required for skeletal muscle atrophy. Science 294, 1704–1708 (2001).
Li, W. et al. Genome-wide and functional annotation of human E3 ubiquitin ligases identifies MULAN, a mitochondrial E3 that regulates the organelle's dynamics and signaling. PLoS ONE 3, e1487 (2008).
Carvalho, P., Goder, V. & Rapoport, T. A. Distinct ubiquitin-ligase complexes define convergent pathways for the degradation of ER proteins. Cell 126, 361–373 (2006). Describes the identification of E3 ligase complexes involved in spatial ubiquitylation and degradation of proteins at the ER.
Biederer, T., Volkwein, C. & Sommer, T. Role of Cue1p in ubiquitination and degradation at the ER surface. Science 278, 1806–1809 (1997).
Ravid, T. & Hochstrasser, M. Autoregulation of an E2 enzyme by ubiquitin-chain assembly on its catalytic residue. Nature Cell Biol. 9, 422–427 (2007).
Neutzner, A., Benard, G., Youle, R. J. & Karbowski, M. Role of the ubiquitin conjugation system in the maintenance of mitochondrial homeostasis. Ann. N. Y. Acad. Sci. 1147, 242–253 (2008).
Cohen, M. M., Leboucher, G. P., Livnat-Levanon, N., Glickman, M. H. & Weissman, A. M. Ubiquitin- proteasome-dependent degradation of a mitofusin, a critical regulator of mitochondrial fusion. Mol. Biol. Cell 19, 2457–2464 (2008).
Yonashiro, R. et al. A novel mitochondrial ubiquitin ligase plays a critical role in mitochondrial dynamics. EMBO J. 25, 3618–3626 (2006).
Zhang, B. et al. GIDE is a mitochondrial E3 ubiquitin ligase that induces apoptosis and slows growth. Cell Res. 18, 900–910 (2008).
Plafker, S. M., Plafker, K. S., Weissman, A. M. & Macara, I. G. Ubiquitin charging of human class III ubiquitin-conjugating enzymes triggers their nuclear import. J. Cell Biol. 167, 649–659 (2004).
Jackson, S. P. & Bartek, J. The DNA-damage response in human biology and disease. Nature 461, 1071–1078 (2009).
Huang, J. et al. RAD18 transmits DNA damage signalling to elicit homologous recombination repair. Nature Cell Biol. 11, 592–603 (2009).
Bekker-Jensen, S. et al. HERC2 coordinates ubiquitin-dependent assembly of DNA repair factors on damaged chromosomes. Nature Cell Biol. 12, 80–86 (2010).
Sowa, M. E., Bennett, E. J., Gygi, S. P. & Harper, J. W. Defining the human deubiquitinating enzyme interaction landscape. Cell 138, 389–403 (2009). Systematic proteomic analysis of DUB interaction networks.
Nijman, S. M. et al. A genomic and functional inventory of deubiquitinating enzymes. Cell 123, 773–786 (2005).
Nakamura, N. & Hirose, S. Regulation of mitochondrial morphology by USP30, a deubiquitinating enzyme present in the mitochondrial outer membrane. Mol. Biol. Cell 19, 1903–1911 (2008).
Hassink, G. C. et al. The ER-resident ubiquitin-specific protease 19 participates in the UPR and rescues ERAD substrates. EMBO Rep. 10, 755–761 (2009).
Zhong, X. & Pittman, R. N. Ataxin-3 binds VCP/p97 and regulates retrotranslocation of ERAD substrates. Hum. Mol. Genet. 15, 2409–2420 (2006).
Nicassio, F. et al. Human USP3 is a chromatin modifier required for S phase progression and genome stability. Curr. Biol. 17, 1972–1977 (2007).
Joo, H. Y. et al. Regulation of cell cycle progression and gene expression by H2A deubiquitination. Nature 449, 1068–1072 (2007).
Sobhian, B. et al. RAP80 targets BRCA1 to specific ubiquitin structures at DNA damage sites. Science 316, 1198–1202 (2007).
Shao, G. et al. The Rap80-BRCC36 de-ubiquitinating enzyme complex antagonizes RNF8-Ubc13-dependent ubiquitination events at DNA double strand breaks. Proc. Natl Acad. Sci. USA 106, 3166–3171 (2009). References 43 and 44 show that spatial control of ubiquitin-editing mechanisms at the sites of DSBs is regulated by the ubiquitin-binding protein RAP80.
Huang, T. T. et al. Regulation of monoubiquitinated PCNA by DUB autocleavage. Nature Cell Biol. 8, 339–347 (2006).
Nijman, S. M. et al. The deubiquitinating enzyme USP1 regulates the Fanconi anemia pathway. Mol. Cell 17, 331–339 (2005).
Sims, A. E. et al. FANCI is a second monoubiquitinated member of the Fanconi anemia pathway. Nature Struct. Mol. Biol. 14, 564–567 (2007).
Yao, T. et al. Distinct modes of regulation of the Uch37 deubiquitinating enzyme in the proteasome and in the Ino80 chromatin-remodeling complex. Mol. Cell 31, 909–917 (2008).
Nakada, S. et al. Non-canonical inhibition of DNA damage-dependent ubiquitination by OTUB1. Nature 466, 941–946 (2010).
Yao, T. et al. Proteasome recruitment and activation of the Uch37 deubiquitinating enzyme by Adrm1. Nature Cell Biol. 8, 994–1002 (2006).
Hamazaki, J. et al. A novel proteasome interacting protein recruits the deubiquitinating enzyme UCH37 to 26S proteasomes. EMBO J. 25, 4524–36 (2006).
Winget, J. M. & Mayor, T. The diversity of ubiquitin recognition: hot spots and varied specificity. Mol. Cell 38, 627–635 (2010).
Mukhopadhyay, D. & Riezman, H. Proteasome-independent functions of ubiquitin in endocytosis and signaling. Science 315, 201–205 (2007).
Bergink, S. & Jentsch, S. Principles of ubiquitin and SUMO modifications in DNA repair. Nature 458, 461–467 (2009).
Bienko, M. et al. Ubiquitin-binding domains in Y-family polymerases regulate translesion synthesis. Science 310, 1821–1824 (2005).
Pinato, S., Gatti, M., Scandiuzzi, C., Confalonieri, S. & Penengo, L. UMI, a novel RNF168 ubiquitin binding domain involved in the DNA damage signaling pathway. Mol. Cell. Biol. 31, 118–126 (2011).
Pinato, S. et al. RNF168, a new RING finger, MIU-containing protein that modifies chromatin by ubiquitination of histones H2A and H2AX. BMC Mol. Biol. 10, 55 (2009).
Bianchi, K. & Meier, P. A tangled web of ubiquitin chains: breaking news in TNF-R1 signaling. Mol. Cell 36, 736–742 (2009).
Hayden, M. S. & Ghosh, S. Shared principles in NF-κB signaling. Cell 132, 344–362 (2008).
Skaug, B., Jiang, X. & Chen, Z. J. The role of ubiquitin in NF-κB regulatory pathways. Annu. Rev. Biochem. 78, 769–796 (2009).
Haas, T. L. et al. Recruitment of the linear ubiquitin chain assembly complex stabilizes the TNF-R1 signaling complex and is required for TNF-mediated gene induction. Mol. Cell 36, 831–844 (2009). By using a proteomic approach, the authors describe spatial recruitment of the linear ubiquitin ligase complex LUBAC to the activated TNFR complexes.
Rahighi, S. et al. Specific recognition of linear ubiquitin chains by NEMO is important for NF-κB activation. Cell 136, 1098–1109 (2009). Describes the structure of the UBAN domain of NEMO in complex with linear diubiquitin, which explains the compartment-based specificity in activation of the IKK complex.
Tokunaga, F. et al. Involvement of linear polyubiquitylation of NEMO in NF-κB activation. Nature Cell Biol. 11, 123–132 (2009).
Ikeda, F. et al. SHARPIN forms a linear ubiquitin ligase complex regulating NF-κB activity and apoptosis Nature 471, 637–641 (2011).
Gerlach, B. et al. Linear ubiquitination prevents inflammation and regulates immune signaling. Nature 471, 591–596 (2011).
Tokunaga, F. et al. Sharpin is a component of the NF-κB activating linear ubiquitin chain assembly complex. Nature 471, 633–636 (2011). References 63–66 provide evidence for the spatiotemporal control of LUBAC (SHARPIN–HOIP–HOIL1)in ubiquitylating NEMO and mediating NF-κB activation in vivo . Deficiency in LUBAC functions leads to multi-organ inflammation and immune system defects.
Dynek, J. N. et al. c-IAP1 and UbcH5 promote K11-linked polyubiquitination of RIP1 in TNF signalling. EMBO J. 29, 4198–4209 (2010).
Ivins, F. J. et al. NEMO oligomerization and its ubiquitin-binding properties. Biochem. J. 421, 243–251 (2009).
Lo, Y. C. et al. Structural basis for recognition of diubiquitins by NEMO. Mol. Cell 33, 602–615 (2009).
Laplantine, E. et al. NEMO specifically recognizes K63-linked poly-ubiquitin chains through a new bipartite ubiquitin-binding domain. EMBO J. 28, 2885–2895 (2009).
Harhaj, E. W. & Dixit, V. M. Deubiquitinases in the regulation of NF-κB signaling. Cell Res. 21, 22–39 (2011).
Komander, D. et al. Molecular discrimination of structurally equivalent Lys 63-linked and linear polyubiquitin chains. EMBO Rep. 10, 466–473 (2009).
Reiley, W., Zhang, M., Wu, X., Granger, E. & Sun, S. C. Regulation of the deubiquitinating enzyme CYLD by IκB kinase γ-dependent phosphorylation. Mol. Cell. Biol. 25, 3886–3895 (2005).
Iha, H. et al. Inflammatory cardiac valvulitis in TAX1BP1-deficient mice through selective NF-κB activation. EMBO J. 27, 629–41 (2008).
Wertz, I. E. et al. De-ubiquitination and ubiquitin ligase domains of A20 downregulate NF-κB signalling. Nature 430, 694–699 (2004).
Lin, S. C. et al. Molecular basis for the unique deubiquitinating activity of the NF-κB inhibitor A20. J. Mol. Biol. 376, 526–540 (2008).
Shembade, N., Ma, A. & Harhaj, E. W. Inhibition of NF-κB signaling by A20 through disruption of ubiquitin enzyme complexes. Science 327, 1135–1139 (2010).
Haglund, K. & Dikic, I. Ubiquitylation and cell signaling. EMBO J. 24, 3353–3359 (2005).
Polo, S. & Di Fiore, P. P. Endocytosis conducts the cell signaling orchestra. Cell 124, 897–900 (2006).
Belgareh-Touze, N. et al. Versatile role of the yeast ubiquitin ligase Rsp5p in intracellular trafficking. Biochem. Soc. Trans. 36, 791–796 (2008).
Saksena, S., Sun, J., Chu, T. & Emr, S. D. ESCRTing proteins in the endocytic pathway. Trends Biochem. Sci. 32, 561–573 (2007).
Hurley, J. H. & Hanson, P. I. Membrane budding and scission by the ESCRT machinery: it's all in the neck. Nature Rev. Mol. Cell Biol. 11, 556–566 (2010).
Shields, S. B. et al. ESCRT ubiquitin-binding domains function cooperatively during MVB cargo sorting. J. Cell Biol. 185, 213–224 (2009).
Clague, M. J. & Urbe, S. Endocytosis: the DUB version. Trends Cell Biol. 16, 551–559 (2006).
Kee, Y., Lyon, N. & Huibregtse, J. M. The Rsp5 ubiquitin ligase is coupled to and antagonized by the Ubp2 deubiquitinating enzyme. EMBO J. 24, 2414–2424 (2005).
Ren, J., Kee, Y., Huibregtse, J. M. & Piper, R. C. Hse1, a component of the yeast Hrs-STAM ubiquitin-sorting complex, associates with ubiquitin peptidases and a ligase to control sorting efficiency into multivesicular bodies. Mol. Biol. Cell 18, 324–335 (2007).
Lin, C. H., MacGurn, J. A., Chu, T., Stefan, C. J. & Emr, S. D. Arrestin-related ubiquitin-ligase adaptors regulate endocytosis and protein turnover at the cell surface. Cell 135, 714–725 (2008). Describes spatial organization of the endocytic complexes at the cell surface by the family of arrestin-related proteins.
Xu, P. et al. Quantitative proteomics reveals the function of unconventional ubiquitin chains in proteasomal degradation. Cell 137, 133–145 (2009). Quantitative proteomics identifies all non-Lys63 ubiquitin chains in proteasomal degradation.
Husnjak, K. et al. Proteasome subunit Rpn13 is a novel ubiquitin receptor. Nature 453, 481–488 (2008).
Schreiner, P. et al. Ubiquitin docking at the proteasome through a novel pleckstrin-homology domain interaction. Nature 453, 548–552 (2008). References 89 and 90 identify a new UBD present in Rpn13, a ubiquitin receptor at the proteasome, that is implicated in binding and recruiting ubiquitylated proteins for degradation.
Glickman, M. H. & Raveh, D. Proteasome plasticity. FEBS Lett. 579, 3214–3223 (2005).
Grabbe, C. & Dikic, I. Functional roles of ubiquitin-like domain (ULD) and ubiquitin-binding domain (UBD) containing proteins. Chem. Rev. 109, 1481–1494 (2009).
Richly, H. et al. A series of ubiquitin binding factors connects CDC48/p97 to substrate multiubiquitylation and proteasomal targeting. Cell 120, 73–84 (2005).
Lee, M. J., Lee, B. H., Hanna, J., King, R. W. & Finley, D. Trimming of ubiquitin chains by proteasome-associated deubiquitinating enzymes. Mol. Cell. Proteomics 7 Sept 2010 (doi:10.1074/mcp.R110.003871).
Crosas, B. et al. Ubiquitin chains are remodeled at the proteasome by opposing ubiquitin ligase and deubiquitinating activities. Cell 127, 1401–1413 (2006). Provides evidence for local editing of ubiquitin chains by opposing activities of E3 ligases and DUBs at the proteasome.
Kraft, C., Peter, M. & Hofmann, K. Selective autophagy: ubiquitin-mediated recognition and beyond. Nature Cell Biol. 12, 836–841 (2010).
Haupt, Y., Maya, R., Kazaz, A. & Oren, M. Mdm2 promotes the rapid degradation of p53. Nature 387, 296–299 (1997).
Lee, J. T. & Gu, W. The multiple levels of regulation by p53 ubiquitination. Cell Death Differ. 17, 86–92 (2010).
Li, M. et al. Mono- versus polyubiquitination: differential control of p53 fate by Mdm2. Science 302, 1972–1975 (2003).
Stommel, J. M. et al. A leucine-rich nuclear export signal in the p53 tetramerization domain: regulation of subcellular localization and p53 activity by NES masking. EMBO J. 18, 1660–1672 (1999).
Lohrum, M. A., Woods, D. B., Ludwig, R. L., Balint, E. & Vousden, K. H. C-terminal ubiquitination of p53 contributes to nuclear export. Mol. Cell. Biol. 21, 8521–8532 (2001).
Mihara, M. et al. p53 has a direct apoptogenic role at the mitochondria. Mol. Cell 11, 577–590 (2003).
Tasdemir, E. et al. Regulation of autophagy by cytoplasmic p53. Nature Cell Biol. 10, 676–687 (2008).
Yamasaki, S. et al. Cytoplasmic destruction of p53 by the endoplasmic reticulum-resident ubiquitin ligase 'Synoviolin'. EMBO J. 26, 113–122 (2007).
Cajigas, I. J., Will, T. & Schuman, E. M. Protein homeostasis and synaptic plasticity. EMBO J. 29, 2746–2752 (2010).
Ehlers, M. D. Activity level controls postsynaptic composition and signaling via the ubiquitin-proteasome system. Nature Neurosci. 6, 231–242 (2003).
Konishi, Y., Stegmuller, J., Matsuda, T., Bonni, S. & Bonni, A. Cdh1-APC controls axonal growth and patterning in the mammalian brain. Science 303, 1026–1030 (2004).
Yang, Y. et al. A Cdc20-APC ubiquitin signaling pathway regulates presynaptic differentiation. Science 326, 575–578 (2009). Shows that the E3 ubiquitin ligase APCCDC20 is important for proper synapse formation in the developing neurons in the brain, possibly through the degradation of brain-enriched transcription factor NEUROD2.
Yan, D., Guo, L. & Wang, Y. Requirement of dendritic Akt degradation by the ubiquitin-proteasome system for neuronal polarity. J. Cell Biol. 174, 415–424 (2006).
Campbell, D. S. & Holt, C. E. Chemotropic responses of retinal growth cones mediated by rapid local protein synthesis and degradation. Neuron 32, 1013–1026 (2001).
Patrick, G. N., Bingol, B., Weld, H. A. & Schuman, E. M. Ubiquitin-mediated proteasome activity is required for agonist-induced endocytosis of GluRs. Curr. Biol. 13, 2073–2081 (2003).
Simpson, J. H., Kidd, T., Bland, K. S. & Goodman, C. S. Short-range and long-range guidance by Slit and its Robo receptors. Robo and Robo2 play distinct roles in midline guidance. Neuron 28, 753–766 (2000).
Myat, A. et al. Drosophila Nedd4, a ubiquitin ligase, is recruited by Commissureless to control cell surface levels of the roundabout receptor. Neuron 35, 447–459 (2002).
Keleman, K. et al. Comm sorts Robo to control axon guidance at the Drosophila midline. Cell 110, 415–427 (2002).
Speese, S. D., Trotta, N., Rodesch, C. K., Aravamudan, B. & Broadie, K. The ubiquitin proteasome system acutely regulates presynaptic protein turnover and synaptic efficacy. Curr. Biol. 13, 899–910 (2003).
Yao, I. et al. SCRAPPER-dependent ubiquitination of active zone protein RIM1 regulates synaptic vesicle release. Cell 130, 943–957 (2007). Reports the identification of the synaptic E3 ligase SCRAPPER as a regulator of proteasome-mediated degradation of RIM1 required for synaptic tuning.
Juo, P. & Kaplan, J. M. The anaphase-promoting complex regulates the abundance of GLR-1 glutamate receptors in the ventral nerve cord of C. elegans. Curr. Biol. 14, 2057–2062 (2004).
Schaefer, H. & Rongo, C. KEL-8 is a substrate receptor for CUL3-dependent ubiquitin ligase that regulates synaptic glutamate receptor turnover. Mol. Biol. Cell 17, 1250–1260 (2006).
Dreier, L., Burbea, M. & Kaplan, J. M. LIN-23-mediated degradation of β-catenin regulates the abundance of GLR-1 glutamate receptors in the ventral nerve cord of C. elegans. Neuron 46, 51–64 (2005).
O'Connor, M. B., Umulis, D., Othmer, H. G. & Blair, S. S. Shaping BMP morphogen gradients in the Drosophila embryo and pupal wing. Development 133, 183–193 (2006).
Podos, S. D., Hanson, K. K., Wang, Y. C. & Ferguson, E. L. The DSmurf ubiquitin-protein ligase restricts BMP signaling spatially and temporally during Drosophila embryogenesis. Dev. Cell 1, 567–578 (2001).
Miles, W. O. et al. Medea SUMOylation restricts the signaling range of the Dpp morphogen in the Drosophila embryo. Genes Dev. 22, 2578–2590 (2008). References 121 and 122 describe the role of ubiquitylation and sumoylation in the regulation of spatial and temporal organization of signalling pathways during embryo development.
Slack, C., Overton, P. M., Tuxworth, R. I. & Chia, W. Asymmetric localisation of Miranda and its cargo proteins during neuroblast division requires the anaphase-promoting complex/cyclosome. Development 134, 3781–3787 (2007).
Silies, M. & Klambt, C. APC/CFzr/Cdh1-dependent regulation of cell adhesion controls glial migration in the Drosophila PNS. Nature Neurosci. 13, 1357–1364 (2010).
Diestel, S., Schaefer, D., Cremer, H. & Schmitz, B. NCAM is ubiquitylated, endocytosed and recycled in neurons. J. Cell Sci. 120, 4035–4049 (2007).
Arama, E., Bader, M., Rieckhof, G. E. & Steller, H. A ubiquitin ligase complex regulates caspase activation during sperm differentiation in Drosophila. PLoS Biol. 5, e251 (2007).
Kaplan, Y., Gibbs-Bar, L., Kalifa, Y., Feinstein-Rotkopf, Y. & Arama, E. Gradients of a ubiquitin E3 ligase inhibitor and a caspase inhibitor determine differentiation or death in spermatids. Dev. Cell 19, 160–173 (2010).
Lipinszki, Z. et al. Developmental-stage-specific regulation of the polyubiquitin receptors in Drosophila melanogaster. J. Cell Sci. 122, 3083–3092 (2009).
Vernace, V. A., Arnaud, L., Schmidt-Glenewinkel, T. & Figueiredo-Pereira, M. E. Aging perturbs 26S proteasome assembly in Drosophila melanogaster. FASEB J. 21, 2672–82 (2007).
Mukai, A. et al. Balanced ubiquitylation and deubiquitylation of Frizzled regulate cellular responsiveness to Wg/Wnt. EMBO J. 29, 2114–2125 (2010).
Tanaka, A. et al. Proteasome and p97 mediate mitophagy and degradation of mitofusins induced by Parkin. J. Cell Biol. 191, 1367–1380 (2010).
Geisler, S. et al. PINK1/Parkin-mediated mitophagy is dependent on VDAC1 and p62/SQSTM1. Nature Cell Biol. 12, 119–131 (2010).
Dixit, E. et al. Peroxisomes are signaling platforms for antiviral innate immunity. Cell 141, 668–681 (2010). Describes the local organization of signalling complexes at the peroxisomal membranes that are essential for antiviral responses.
Cohen, P. & Tcherpakov, M. Will the ubiquitin system furnish as many drug targets as protein kinases. Cell 143, 686–693 (2010).
Hoeller, D. & Dikic, I. Targeting the ubiquitin system in cancer therapy. Nature 458, 438–444 (2009).
Varshavsky, A. The ubiquitin system. Trends Biochem. Sci. 22, 383–387 (1997).
Rabut, G. & Peter, M. Function and regulation of protein neddylation. 'Protein modifications: beyond the usual suspects' review series. EMBO Rep. 9, 969–976 (2008).
Meulmeester, E., Kunze, M., Hsiao, H. H., Urlaub, H. & Melchior, F. Mechanism and consequences for paralog-specific sumoylation of ubiquitin-specific protease 25. Mol. Cell 30, 610–619 (2008).
Gallagher, E., Gao, M., Liu, Y. C. & Karin, M. Activation of the E3 ubiquitin ligase Itch through a phosphorylation-induced conformational change. Proc. Natl Acad. Sci. USA 103, 1717–1722 (2006).
Pastori, V. et al. CK2 and GSK3 phosphorylation on S29 controls wild-type ATXN3 nuclear uptake. Biochim. Biophys. Acta 1802, 583–592 (2010).
Varshavsky, A. The N-end rule. Cell 69, 725–735 (1992).
Hwang, C. S., Shemorry, A. & Varshavsky, A. N-terminal acetylation of cellular proteins creates specific degradation signals. Science 327, 973–977 (2010).
Lallemand-Breitenbach, V. et al. Arsenic degrades PML or PML-RARα through a SUMO-triggered RNF4/ubiquitin-mediated pathway. Nature Cell Biol. 10, 547–555 (2008).
Tatham, M. H. et al. RNF4 is a poly-SUMO-specific E3 ubiquitin ligase required for arsenic-induced PML degradation. Nature Cell Biol. 10, 538–546 (2008). References 143 and 144 provide evidence for regulated ubiquitylation of the promyelocytic leukaemia–retinoic acid receptor-α complex upon recruitment of E3 ligase RNF4 to SUMO chains. This pathway is used as an anti-cancer treatment mechanism through treatment with arsenic trioxide.
Acknowledgements
We apologize to all scientists whose important contributions were not referenced in this Review owing to limitations in the number of references. We are grateful to C. Joazeiro and H. Walczak for comments and discussions. C.G. is supported by the Swedish Foundation for Strategic Research (SSF). Research in the I.D. laboratory is supported by the Deutsche Forschungsgemeinschaft, the Cluster of Excellence 'Macromolecular Complexes' of the Goethe University Frankfurt (EXC115) and a European Research Council Advanced Grant.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Related links
Glossary
- RING
-
A zinc-binding protein–protein interaction motif, found in RING-type ubiquitin E3 ligases, that binds to the E2 ubiquitin thioester and thereby promotes ubiquitin transfer to substrate proteins.
- ER-associated degradation
-
A protein quality control pathway (mediated by p97–VCP–CDC48) in which misfolded or regulated proteins residing in the endoplasmic reticulum (ER) are translocated into the cytosol for proteasomal degradation.
- Ubiquitin-like molecule
-
A protein that structurally resembles ubiquitin and uses its own E1, E2 and E3 enzymes to conjugate to target proteins and to modify their properties. Examples are the small ubiquitin-like modifiers (SUMOs), NEDD8, ISG15, Fat10 and autophagy-related 8.
- N-end rule
-
A biological principle that relates the half-life of a cellular protein to the identity of its amino-terminal residue, in which N-terminal amino acids are recognized by specific E3 ubiquitin ligases (N-recognins).
- Breast cancer type 1 susceptibility
-
(BRCA1). An E3 ligase that catalyses multiple types of ubiquitin signals. It is often found in a heterodimeric RING complex with BRCA1-associated RING domain 1 and has an important role in homologous recombination repair of DNA double-strand breaks.
- Ubiquitin-interacting motif
-
An α-helical ubiquitin-binding domain that binds to the conserved hydrophobic patch that is centred around Ile44 in ubiquitin with an affinity in the range of ∼100–400M.
- HECT
-
One of the major classes of ubiquitin ligases. HECT E3 ligases contain a domain with a catalytic Cys residue that forms a thioester intermediate during ubiquitin transfer to the substrate protein.
- Proliferating cell nuclear antigen
-
(PCNA). A ring-shaped molecule encircling DNA. It slides bidirectionally along DNA to constitutively monitor genomic integrity. Following DNA damage, ubiquitylation of PCNA is essential for the recruitment of damage-tolerant DNA polymerases, allowing translesion synthesis.
- F-box
-
A protein module of ∼50 residues that is involved in mediating protein–protein interactions. F–box proteins commonly act as substrate recognition subunits in cullin–RING ubiquitin ligases.
- Receptor Tyr kinase
-
A membrane-bound protein Tyr kinase that often functions as a receptor for secreted hormones, growth factors and cytokines.
- Diubiquitin motif
-
A double-sided ubiquitin-binding domain, first identified in hepatocyte growth factor-regulated Tyr kinase substrate (HRS), that can simultaneously bind two ubiquitin moieties, both of which are required for the endocytic sorting function of HRS.
- GRAM-like ubiquitin-binding in EAP45
-
A ubiquitin-binding domain that folds into a split pleckstrin homology domain with a non-canonical lipid-binding pocket that interacts with phosphatidylinositol-3-phosphate.
- Multivesicular body
-
(MVB). An intermediate structure in the endosomal pathway that is formed when membrane portions bud into the lumen of late endosomes, forming intralumenal vesicles. MVB sorting is an essential event for the degradation of internalized cell surface proteins in the lysosome.
- Ubiquitin-associated domain
-
A short (∼40 amino acids) sequence motif, first found in proteins associated with the ubiquitylation pathway, that mediates polyubiquitin binding.
- Ubiquitin-like domain
-
(UBL domain). A modular protein domain present in multiple cellular proteins. UBL domains structurally resemble ubiquitin by folding into the ubiquitin β-grasp superfold and can in general be recognized by ubiquitin-binding domains.
- JAMM
-
(JAB1/MPN/MOV34 metalloenzyme). A family of zinc metalloprotease deubiquitylating enzymes.
- Anaphase-promoting complex
-
A multifunctional ubiquitin ligase that, by pairing with its co-activators CDC20 and CDH1, specifically targets cell cycle proteins (among others) for degradation.
- Cullin–RING ubiquitin ligase
-
(CRL). A member of a large family of multisubunit E3 ligases, commonly comprising a cullin scaffold, a catalytic RING subunit, a substrate-recognition subunit (SRS), and, for most CRLs, an adaptor subunit linking the SRS to the complex.
- Postsynaptic density
-
(PSD). A structure in the postsynaptic membrane in which LGlu neurotransmitter receptors are accumulated together with multiple adhesion, scaffold, cytoskeletal and signalling molecules. PSDs organize the postsynaptic signalling machinery, control synaptic plasticity and maintain synaptic homeostasis.
Rights and permissions
About this article
Cite this article
Grabbe, C., Husnjak, K. & Dikic, I. The spatial and temporal organization of ubiquitin networks. Nat Rev Mol Cell Biol 12, 295–307 (2011). https://doi.org/10.1038/nrm3099
Published:
Issue Date:
DOI: https://doi.org/10.1038/nrm3099
This article is cited by
-
The therapeutic potential of targeting regulated non-apoptotic cell death
Nature Reviews Drug Discovery (2023)
-
Ubiquitin ligase enzymes and de-ubiquitinating enzymes regulate innate immunity in the TLR, NLR, RLR, and cGAS-STING pathways
Immunologic Research (2023)
-
Semantic clustering analysis of E3-ubiquitin ligases in gastrointestinal tract defines genes ontology clusters with tissue expression patterns
BMC Gastroenterology (2022)
-
USP8 inhibition reshapes an inflamed tumor microenvironment that potentiates the immunotherapy
Nature Communications (2022)
-
β-Klotho promotes glycolysis and glucose-stimulated insulin secretion via GP130
Nature Metabolism (2022)