Key Points
-
Nucleic-acid aptamers can be isolated robotically in vitro to provide novel molecular recognition tools for research. They can be developed for use in various applications, including diagnostics and therapeutics.
-
Most molecular targets can be used to isolate tightly binding aptamers, potentially extending aptamer application to most molecular sciences.
-
Analysis of aptamers has yielded insights into the evolution of natural protein/ligand–nucleic-acid complexes.
-
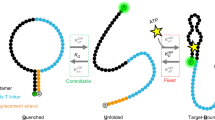
Small-molecule ligand binding to RNA aptamers often induces conformational changes that can be used to develop novel diagnostic reagents.
-
Nature has made use of these properties to evolve riboswitches — sequence elements that directly regulate gene expression at the level of mRNA in response to ligand binding and temperature changes.
-
Clinically effective aptamer drugs are now entering use and many more are in clinical trials.
-
Research has identified a large number of potential anti-viral aptamers, including those targeted against HIV, hepatitis C virus and influenza virus.
Abstract
Nucleic-acid aptamers have the molecular recognition properties of antibodies, and can be isolated robotically for high-throughput applications in diagnostics, research and therapeutics. Unlike antibodies, however, they can be chemically derivatized easily to extend their lifetimes in biological fluids and their bioavailability in animals. The first aptamer-based clinical drugs have recently entered service. Meanwhile, active research programmes have identified a wide range of anti-viral aptamers that could form the basis for future therapeutics.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
References
Kruger, K. et al. Self-splicing RNA: autoexcision and autocyclization of the ribosomal RNA intervening sequence of tetrahymena. Cell 31, 147–157 (1982).
Stark, B. C., Kole, R., Bowman, E. J. & Altman, S. Ribonuclease P: an enzyme with an essential RNA component. Proc. Natl Acad. Sci. USA 75, 3712–3721 (1978).
Robertson, D. L. & Joyce, G. F. Selection in vitro of an RNA enzyme that specifically cleaves single-stranded DNA. Nature Struct. Biol. 344, 467–468 (1990).
Tuerk, C. & Gold, L. Systematic evolution of ligands by exponential enrichment: RNA ligands to bacteriophage T4 DNA-polymerase. Science 249, 505–510 (1990). This paper describes the first use of the term SELEX and explains the basics of the process.
Ellington, A. D. & Szostak, J. W. In vitro selection of RNA molecules that bind specific ligands. Nature 346, 818–822 (1990).
Wilson, C. & Szostak, J. W. In vitro evolution of a self-alkylatlng ribozyme. Nature 374, 766–767 (1995).
Seelig, B. & Jaschke, A. A small catalytic RNA motif with Diels–Alderase activity. Chem. Biol. 6, 167–176 (1999).
Gilbert, W. The RNA world. Nature 319, 818 (1986).
Cox, J. C. et al. Automated selection of aptamers against protein targets translated in vitro: from gene to aptamer. Nucleic Acids Res. 30, e108 (2002).
Cox, J. C., Rudolph, P. & Ellington, A. D. Automated RNA selection. Biotechnol. Prog. 14, 845–850 (1998).
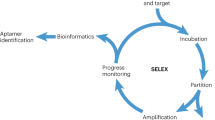
Ellington, A. D., Cox, J. C., Lee, J. F. & Collett, J. R. in The RNA World 3rd edn (eds Gesteland, R. F., Cech, T. R. & Atkins, J. F.) 683–719 (Cold Spring Harbor Press, New York, 2006). This chapter explains the basic considerations of all aptamer isolation experiments from initial pool diversity and library size to high-throughput techniques. Applications of aptamers are also briefly discussed.
Mandal, M. & Breaker, R. R. Gene regulation by riboswitches. Nature Rev. Mol. Cell Biol. 5, 451–463 (2004).
Tucker, B. J. & Breaker, R. R. Riboswitches as versatile gene control elements. Curr. Opin. Struct. Biol. 15, 342–348 (2005).
Schachat, A. P. New treatments for age-related macular degeneration. Ophthalmology 112, 531–532 (2005).
Lee, J. H. et al. A therapeutic aptamer inhibits angiogenesis by specifically targeting the heparin binding domain of VEGF165. Proc. Natl Acad. Sci. USA 102, 18902–18907 (2005).
Ng, E. W. M. et al. Pegaptanib, a targeted anti-VEGF aptamer for ocular vascular disease. Nature Rev. Drug Discov. 5, 123–132 (2006). This paper describes the various steps in creating the first clinically available therapeutic aptamer, from initial isolation to subsequent development.
Shtatland, T. et al. Interactions of Escherichia coli RNA with bacteriophage MS2 coat protein: genomic SELEX. Nucleic Acids Res. 28, e93 (2000).
Hirao, I., Spingola, M., Peabody, D. & Ellington, A. D. The limits of specificity: an experimental analysis with RNA aptamers to MS2 coat protein variants. Mol. Divers. 4, 75–89 (1999).
Rowsell, S. et al. Crystal structures of a series of RNA aptamers complexed to the same protein target. Nature Struct. Biol. 5, 970–975 (1998).
Convery, M. A. et al. The crystal structure of an RNA aptamer protein complex at 2.8Å resolution. Nature Struct. Biol. 5, 133–139 (1998).
He, Y.-Y., Stockley, P. G. & Gold, L. In vitro evolution of the DNA binding sites of Escherichia coli methionine repressor, MetJ. J. Mol. Biol. 255, 55–66 (1996).
Wang, Y. & Rando, R. R. Specific binding of aminoglycoside antibiotics to RNA. Chem. Biol. 2, 281–290 (1995).
Wallace, S. T. & Schroeder, R. In vitro selection and characterization of RNAs with high affinity to antibiotics. Meth. Enzymol. 318, 214–229 (2000).
Jiang, L. & Patel, D. J. Solution structure of the tobramycin-RNA aptamer complex. Nature Struct. Biol. 5, 769–774 (1998).
Tereshko, V., Skripkin, E. & Patel, D. J. Encapsulating streptomycin within a small 40-mer RNA. Chem. Biol. 10, 175–187 (2003).
Serganov, A. et al. Structural basis for Diels–Alder ribozyme-catalyzed carbon–carbon bond formation. Nature Struct. Mol. Biol. 12, 218–224 (2005).
Khati, M. et al. Neutralization of infectivity of diverse R5 clinical isolates of human immunodeficiency virus type 1 by gp120-binding 2′ F-RNA aptamers. J. Virol. 77, 12692–12698 (2003).
Misono, T. S. & Kumar, P. K. R. Selection of RNA aptamers against human influenza virus hemagglutinin using surface plasmon resonance. Anal. Biochem. 342, 312–317 (2005).
Berezovski, M. et al. Nonequilibrium capillary electrophoresis of equilibrium mixtures: a universal tool for development of aptamers. J. Am. Chem. Soc. 127, 3165–3171 (2005).
Pieken, W. A., Olsen, D. B., Benseler, F., Aurup, H. & Eckstein, F. Kinetic characterisation of ribonuclease-resistant hammerhead ribozymes. Science 253, 314–316 (1991).
Beigelman, L. et al. Chemical modification of hammerhead ribozymes. Catalytic activity and nuclease resistance. J. Biol. Chem. 270, 25702–25708 (1995). This paper shows how incorporation of modified nucleotides can dramatically increase the half-lives of aptamers.
Chelliserrykattil, J. & Ellington, A. D. Evolution of a T7 RNA polymerase variant that transcribes 2′-O-methyl RNA. Nature Biotechnol. 22, 1155–1160 (2004).
Kato, Y. et al. New NTP analogs: the synthesis of 4′-thioUTP and 4′-thioCTP and their utility for SELEX. Nucleic Acids Res. 33, 2942–2951 (2005).
Klussmann, S., Nolte, A., Bald, R., Erdmann, V. A. & Fürste, J. P. Mirror-image RNA that binds D-adenosine. Nature Biotechnol. 14, 1112–1115 (1996). This paper outlines an alternative approach to generate nuclease-resistant aptamers that relies on the use of chemical symmetry to create aptamers based on L -ribose.
Osborne, S. E., Völker, J., Stevens, S. Y., Breslauer, K. J. & Glick, G. D. Design, synthesis and analysis of disulphide cross-linked DNA duplexes. J. Am. Chem. Soc. 118, 11993–12003 (1996).
Willis, M. C. et al. Liposome anchored vascular endothelial growth factor aptamers. Bioconjug. Chem. 9, 573–582 (1998).
Ruckman, J. et al. 2′-Fluoropyrimidine RNA-based aptamers to the 165-amino-acid form of the vascular endothelial growth factor (VEGF165). J. Biol. Chem. 273, 20556–20567 (1998).
Boomer, R. M. et al. Conjugation to polyethylene glycol polymer promotes aptamer biodistribution to healthy and inflamed tissues. Oligonucleotides 15, 183–195 (2005).
de Smidt, P. C., Doan, T. L., de Falco, S. & van Berkel, T. J. C. Association of antisense oligonucleotides with lipoproteins prolongs the plasma half-life and modifies the tissue distribution. Nucleic Acids Res. 19, 4695–4700 (1991).
Dougan, H. et al. Extending the lifetime of anticoagulant oligodeoxynucleotide aptamers in blood. Nucl. Med. Bio. 27, 289–297 (2000). This paper shows how further chemical modifications to aptamers can affect their biodistribution and availability.
White, W. et al. Generation of species cross-reactive aptamers using 'toggle' SELEX. Mol. Ther. 4, 567–573 (2001).
Vater, A., Jarosch, F., Buchner, K. & Klussmann, S. Short bioactive Spiegelmers to migraine-associated calcitonin gene-related peptide rapidly identified by a novel approach: tailored-SELEX. Nucleic Acids Res. 31, el30 (2003).
Golden, M. C., Collins, B. D., Willis, M. C., Koch, T. H. Diagnostic potential of PhotoSELEX-evolved ssDNA aptamers. J. Biotechnol. 81, 167–178 (2000).
Romig, T. S., Bell, C. & Drolet, D. W. Aptamer affinity chromatography: combinatorial chemistry applied to protein purification. J. Chromat. 731, 275–284 (1999).
Michaud, M. et al. A DNA aptamer as a new target-specific chiral selector for HPLC. J. Am. Chem. Soc. 125, 8672–8679 (2003).
Koita, R. B., Li, L., McGown, L. B. Separation of non-target compounds by DNA aptamers. Anal. Chem. 72, 827–831 (2000).
Hartmuth, K., Vornlocher, H. P. & Luhrmann, R. Tobramycin affinity tag purification of spliceosomes. Meth. Mol. Biol. 257, 47–64 (2004).
Murphy, M. B., Fuller, S. T., Richardson, P. M. & Doyle, S. A. An improved method for the in vitro evolution of aptamers and applications in protein detection and purification. Nucleic Acids Res. 31, e110 (2003).
Seiwert, S. D., Nahreini, T. S., Aigner, S., Ahn, N. G. & Uhlenbeck, O. C. RNA aptamers as pathway-specific MAP kinase inhibitors. Chem. Biol. 7, 833–843 (2000).
Vuyisich, M. & Beal, P. A. Controlling protein activity with ligand-regulated RNA aptamers. Chem. Biol. 9, 907–913 (2002). This paper demonstrates the use of aptamers to regulate protein activity, showing how aptamers can be activated and de-activated at will, making them more useful than conventional diagnostic and therapeutic agents.
Rusconi, C. P. et al. Aptamers as reversible antagonists of coagulation factor IXa. Nature 419, 90–94 (2002).
Heckel, A. & Mayer, G. Light regulation of aptamer activity: an anti-thrombin aptamer with caged thymidine nucleobases. J. Am. Chem. Soc. 127, 822–823 (2005).
Mayer, G., Kröck, L., Mikat, V., Engeser, M. & Heckel, A. Light-induced formation of G-quadruplex DNA secondary structures. Chem. Biochem. 6, 1966–1970 (2005).
Brockstedt, U., Uzaroeska, A., Montpetit, A., Pfau, W. & Labuda, D. In vitro evolution of RNA aptamers recognizing carcinogenic aromatic amines. Biochem. Biophys. Res. Commun. 313, 1004–1008 (2003).
Stojanovic, M. N. & Kolpashchikov, D. M. Modular aptameric sensors. J. Am. Chem. Soc. 126, 9226–9270 (2004).
Brody, E. N. et al. The use of aptamers in large scale arrays for molecular diagnosis. Mol. Diag. 4, 381–387 (1999).
Collett, J. R., Cho, E. J. & Ellington, A. D. Production and processing of aptamer microarrays. Methods 37, 4–15 (2005).
Yamamoto-Fujita, R. & Kumar, P. K. R. Aptamer-derived nucleic acid oligos: applications to develop nucleic acid chips to analyze proteins and small ligands. Anal. Chem. 77 17, 5460–5466 (2005).
Levy, M., Cater, S. F. & Ellington, A. D. Quantum-dot aptamer beacons for the detection of proteins. Chem. Biochem. 6, 2163–2166 (2005).
Liu, J. & Lu, Y. Fast colorimetric sensing of adenosine and cocaine based on a general sensor design involving aptamers and nanoparticles. Angew. Chem. Int. Ed. Engl. 45, 90–94 (2006).
Tu, D., Blaha, G., Moore, P. B. & Steitz, T. A. Structures of MLSBK antibiotics bound to mutated large ribosomal subunits provide a structural explanation for resistance. Cell 121, 257–270 (2005).
Varghese, J. N., Epa, V. C. & Colman, P. M. Three-dimensional structure of the complex of 4-guanidino-Neu5Ac2en and influenza virus neuraminidase. Protein Sci. 4, 1081–1087 (1995).
Sandberg, J. A. et al. Pharmacokinetics and tolerability of an antiangiogenic ribozyme (Angiozyme) in healthy volunteers. J. Clin. Pharmacol. 40, 1462–1469 (2000). This paper outlines the biodistribution, bioavailability and clearance of RNA-based therapeutics.
Dzau, V. J., Mann, M. J., Morishita, R. & Kandea, Y. Fusigenic viral liposome for gene therapy in cardiovascular diseases. Proc. Natl Acad. Sci. USA 93, 11421–11425 (1996).
Usman, N. & Blatt, L. M. Nuclease-resistant synthetic ribozymes: developing a new class of therapeutics. J. Clin. Invest. 106, 1197–1202 (2000).
Chaloin, L., Lehmann, M. J., Sczakiel, G. & Restle, T. Endogenous expression of a high-affinity pseudoknot RNA aptamer suppresses replication of HIV-1. Nucleic Acid Res. 30, 4001–4008 (2002).
Good, P. D. et al. Expression of small, therapeutic RNAs in human cell nuclei. Gene Ther. 4, 45–54 (1997).
Sullenger, B. A., Gallardo, H. F., Ungers, G. E. & Gilboa, E. Over-expression of TAR sequences renders cells resistant to human immunodeficiency virus replication. Cell 63, 601–608 (1990).
Jellinek, D., Green, L. S., Bell, C. & Janjic, N. Inhibition of receptor-binding by high-affinity RNA ligands to vascular endothelial growth-factor. Biochem. 33, 10450–10456 (1994).
Cerchia, L., Hamm, J., Libri, D., Tavitian, B. & Francisics, V. Nucleic acid aptamers in cancer medicine. FEBS lett. 528, 12–16 (2002). This paper is a short review of the use of aptamers in the treatment of cancers.
Ylera, F., Lurz, R., Erdmann, V. A. & Fürste, J. P. Selection of RNA aptamers to the Alzheimer's disease amyloid peptide. Biochem. Biophys. Res. Commun. 290, 1583–1588 (2002).
Sayer, N. M. et al. Structural determinants of conformationally selective, prion-binding aptamers. J. Biol. Chem. 279, 13102–13109 (2004).
Rhie, A. et al. Characterization of 2′-fluoro-RNA aptamers that bind preferentially to disease-associated conformations of prion protein and inhibit conversion. J. Biol. Chem. 278, 39697–39705 (2003).
Charlton, J., Sennello, J. & Smith, D. In vivo imaging of inflammation using an aptamer inhibitor of human neutrophil elastase. Chem. Biol. 4, 809–816 (1997).
Held, D. M., Kissel, J. D., Patterson, J. T., Nickens, D. G. & Burke, D. H. HIV-1 inactivation by nucleic acid aptamers. Front. Biosci. 11, 89–112 (2006). This paper provides a comprehensive review of aptamers targeting various crucial proteins in HIV.
Tuerk, C., MacDougal, S. & Gold, L. RNA pseudoknots that inhibit human immunodeficiency virus type 1 reverse transcriptase. Proc. Natl Acad. Sci. USA 89, 6988–6992 (1992).
Nickens, D. G., Patterson, J. T. & Burke, D. H. Inhibition of HIV-1 reverse transcriptase by RNA aptamers in Escherichia coli. RNA 9, 1029–1033 (2003).
Allen, P., Worland, S. & Gold, L. Isolation of high affinity RNA ligands to HIV-1 integrase from a random pool. Virol. 209, 327–336 (1995).
Dey, A. K., Griffiths, C., Lea, S. M. & James, W. Structural characterization of an anti-gp120 RNA aptamer that neutralizes R5 strains of HIV-1. RNA 11, 873–884 (2005).
Dey, A. K. et al. An aptamer that neutralizes R5 strains of human immunodeficiency virus type 1 blocks gp120-CCR5 interaction. J. Virol. 79, 13806–13810 (2005).
Hwang, B. et al. Isolation of specific and high-affinity RNA aptamers against NS3 helicase domain of hepatitis C virus. RNA 10, 1277–1290 (2004).
Nishikawa, F., Funaji, K., Fukuda, K. & Nishikawa, S. In vitro selection of RNA aptamers against the HCVNS3 helicase domain. Oligonucleotides 14, 114–129 (2004).
Fukuda, K. et al. An RNA ligand inhibits hepatitis C virus NS3 protease and helicase activities. Biochem. Biophys. Res. Commun. 325, 670–675 (2004).
Zhan, L. S., Zhuo, H. L., Wang, H. Z., Peng, J. C. & Wang, Q. L. Screening and characterization of aptamers of hepatitis C virus NS3 helicase. Prog. Biochem. Biophys. 32, 245–250 (2005).
Hwang, B. & Lee, S. W. Analysis of in vivo interaction of HCVNS3 protein and specific RNA aptamer with yeast three-hybrid system. J. Micro. Biotechnol. 15, 660–664 (2005).
Bellecave, P. et al. Selection of DNA aptamers that bind the RNA-dependent RNA polymerase of hepatitis C virus and inhibit viral RNA synthesis in vitro. Oligonucleotides 13, 455–463 (2003).
Romero-Lopez, C., Barroso- del Jesus, A., Puerta-Fernandez, E. & Berzal-Herranz, A. Interfering with hepatitis C virus IRES activity using RNA molecules identified by a novel in vitro selection method. Biol. Chem. 386, 183–190 (2005).
Kikuchi, K. et al. A hepatitis C virus (HCV) internal ribosome entry site (IRES) domain III-IV-targeted aptamer inhibits translation by binding to an apical loop of domain IIId. Nucleic Acids Res. 33, 683–692 (2005).
Wang, J., Jiang, H. & Liu, F. In vitro selection of novel RNA ligands that bind human cytomegalovirus and block viral infection. RNA 6, 571–583 (2000).
Werstuck, G. & Green, M. R. Controlling gene expression in living cells through small molecule-RNA interactions. Science 282, 296–298 (1998).
Davidson, E. A. & Ellington, A. D. Engineering regulatory RNAs. Trends Biotechnol. 23, 109–112 (2005).
Johansson, J. et al. An RNA thermosensor controls expression of virulence genes in Listeria monocytogenes. Cell 110, 551–561 (2002).
Winkler, W. C., Nahvi, A., Roth, A., Collins, J. A. & Breaker, R. R. Control of gene expression by a natural metabolite-responsive ribozyme. Nature 428, 281–286 (2004).
Yen, L. et al. Exogenous control of mammalian gene expression through modulation of RNA self-cleavage. Nature 431, 471–476 (2004).
Thompson, K. M., Syrett, H. A., Knudsen, S. M. & Ellington, A. D. Group I aptazymes as genetic regulatory switches. BMC Biotechnol. 2, 21 (2002).
Isaacs, F. J. et al. Engineered riboregulators enable post-transcriptional control of gene expression. Nature Biotechnol. 22, 841–847 (2004).
Laserson, U., Gan, H. H. & Schlick, T. Predicting candidate genomic sequences that correspond to synthetic functional RNA motifs. Nucleic Acids Res. 33, 6057–6069 (2005).
Suess, B., Fink, B., Berens, C., Stentz, R. & Hillen, W. A theophylline responsive riboswitch based on helix slipping controls gene expression in vivo. Nucleic Acids Res. 32, 1610–1614 (2004).
Desai, S. K. & Gallivan, J. P. Genetic screens and selections for small molecules based on a synthetic riboswitch that activates protein translation. J. Am. Chem. Soc. 126, 13247–13254 (2004).
Serganov, A. et al. Structural basis for discriminative regulation of gene expression by adenine- and guanine-sensing mRNAs. Chem. Biol. 11, 1729–1741 (2004). This paper reports the first crystal structures for purine responsive riboswitches with bound effector bases. The molecular basis of base discrimination and ligand-sensor domain folding are revealed.
Acknowledgements
We would like to thank D. Burke, A. Ellington, M. Famulok and W. James for helpful comments during the preparation of this manuscript and for sharing unpublished or recent work from their laboratories. We thank W. Horn for providing figure 2. Aptamer research in the P.G.S. laboratory is supported by the UK Medical Research Council and the Biotechnology and Biological Sciences Research Council, and by The Wellcome Trust and The Leverhulme Trust.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Related links
Related links
DATABASES
Entrez Genome Project
FURTHER INFORMATION
Glossary
- Macular degeneration
-
A condition in which the light-sensing cells of the macula, which is in the centre of the retina, malfunction and cease to work, leading to reduction or loss of central vision. The disease can be caused by the leakage of newly forming blood vessels into the retina and it is this process that is susceptible to treatment by Macugen.
- Sequence space
-
All the possible sequence combinations in a nucleic-acid library used for SELEX. As there are many such sequences, it is unlikely that all possible combinations of sequence and function have been 'tried' during evolution.
- Surface plasmon resonance
-
(SPR). A technique for monitoring the affinity between molecules in solution (analytes) as they pass across an immobilized target on the SPR sensorchip. In aptamer research, this technique is used to collect slowly dissociating aptamer species that have higher affinity than those that elute early.
- Spiegelmers
-
A term derived from the German word for mirror. These are RNA aptamers synthesized chemically with L-ribose instead of the natural D-ribose and are therefore resistant to nuclease action.
- Enantiomer
-
Two molecular structures that have identical chemical compositions but are non-superimposable in three dimensions — they are mirror images of each other. For amino acids and ribose sugars these are known as the D- and L-forms.
- Capillary electrochromatography
-
A collection of separation techniques, which involve the application of high voltages across buffer-filled capillaries to achieve separations based on a range of different physical properties.
- Pseudoknot
-
A common three-dimensional feature of RNA, in which bases in a single-stranded loop base pair with complementary bases outside that loop. Pseudoknots are commonly used recognition and control elements in vivo but often stabilize selected aptamers.
Rights and permissions
About this article
Cite this article
Bunka, D., Stockley, P. Aptamers come of age – at last. Nat Rev Microbiol 4, 588–596 (2006). https://doi.org/10.1038/nrmicro1458
Issue Date:
DOI: https://doi.org/10.1038/nrmicro1458
This article is cited by
-
An AuNPs-based electrochemical aptasensor for the detection of 25-hydroxy vitamin D3
Analytical Sciences (2024)
-
Advantages of Material Biofunctionalization Using Nucleic Acid Aptamers in Tissue Engineering and Regenerative Medicine
Molecular Biotechnology (2023)
-
Evaluation of penicillin residues in milk by ELISA using aptamer bonded to gold nanoparticles
Gold Bulletin (2022)
-
Glycated albumin precipitation using aptamer conjugated magnetic nanoparticles
Scientific Reports (2020)
-
Dark field microscope-based single nanoparticle identification coupled with statistical analysis for ultrasensitive biotoxin detection in complex sample matrix
Microchimica Acta (2020)