Abstract
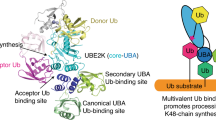
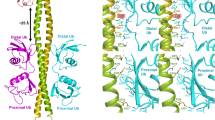
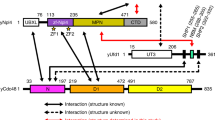
Lys63-linked polyubiquitin chains participate in nonproteolytic signaling pathways, including regulation of DNA damage tolerance and NF-κB activation. E2 enzymes bound to ubiquitin E2 variants (UEV) are vital in these pathways, synthesizing Lys63-linked polyubiquitin chains, but how these complexes achieve specificity for a particular lysine linkage has been unclear. We have determined the crystal structure of an Mms2–Ubc13-ubiquitin (UEV–E2-Ub) covalent intermediate with donor ubiquitin linked to the active site residue of Ubc13. In the structure, the unexpected binding of a donor ubiquitin of one Mms2–Ubc13-Ub complex to the acceptor-binding site of Mms2–Ubc13 in an adjacent complex allows us to visualize at atomic resolution the molecular determinants of acceptor-ubiquitin binding. The structure reveals the key role of Mms2 in allowing selective insertion of Lys63 into the Ubc13 active site and suggests a molecular model for polyubiquitin chain elongation.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$189.00 per year
only $15.75 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
References
Hershko, A. & Ciechanover, A. The ubiquitin system. Annu. Rev. Biochem. 67, 425–479 (1998).
Pickart, C.M. & Fushman, D. Polyubiquitin chains: polymeric protein signals. Curr. Opin. Chem. Biol. 8, 610–616 (2004).
Sun, L. & Chen, Z.J. The novel functions of ubiquitination in signaling. Curr. Opin. Cell Biol. 16, 119–126 (2004).
Ulrich, H.D. Degradation or maintenance: actions of the ubiquitin system on eukaryotic chromatin. Eukaryot. Cell 1, 1–10 (2002).
Pickart, C.M. & Cohen, R.E. Proteasomes and their kin: proteases in the machine age. Nat. Rev. Mol. Cell Biol. 5, 177–187 (2004).
Hicke, L. & Dunn, R. Regulation of membrane protein transport by ubiquitin and ubiquitin-binding proteins. Annu. Rev. Cell Dev. Biol. 19, 141–172 (2003).
Pickart, C.M. & Eddins, M.J. Ubiquitin: structures, functions, mechanisms. Biochim. Biophys. Acta 1695, 55–72 (2004).
Deng, L. et al. Activation of the IκB kinase complex by TRAF6 requires a dimeric ubiquitin-conjugating enzyme complex and a unique polyubiquitin chain. Cell 103, 351–361 (2000).
Hofmann, R.M. & Pickart, C.M. Noncanonical MMS2-encoded ubiquitin-conjugating enzyme functions in assembly of novel polyubiquitin chains for DNA repair. Cell 96, 645–653 (1999).
VanDemark, A.P., Hofmann, R.M., Tsui, C., Pickart, C.M. & Wolberger, C. Molecular insights into polyubiquitin chain assembly: crystal structure of the Mms2/Ubc13 heterodimer. Cell 105, 711–720 (2001).
Moraes, T.F. et al. Crystal structure of the human ubiquitin conjugating enzyme complex, hMms2-hUbc13. Nat. Struct. Biol. 8, 669–673 (2001).
Andersen, P.L. et al. Distinct regulation of Ubc13 functions by the two ubiquitin-conjugating enzyme variants Mms2 and Uev1A. J. Cell Biol. 170, 745–755 (2005).
Ulrich, H.D. The RAD6 pathway: control of DNA damage bypass and mutagenesis by ubiquitin and SUMO. ChemBioChem 6, 1735–1743 (2005).
Chen, Z.J. Ubiquitin signalling in the NF-kappaB pathway. Nat. Cell Biol. 7, 758–765 (2005).
Zhou, H. et al. Bcl10 activates the NF-kappaB pathway through ubiquitination of NEMO. Nature 427, 167–171 (2004).
Hamilton, K.S. et al. Structure of a conjugating enzyme-ubiquitin thiolester intermediate reveals a novel role for the ubiquitin tail. Structure 9, 897–904 (2001).
McKenna, S. et al. Energetics and specificity of interactions within UbUevUbc13 human ubiquitin conjugation complexes. Biochemistry 42, 7922–7930 (2003).
McKenna, S. et al. An NMR-based model of the ubiquitin-bound human ubiquitin conjugation complex Mms2Ubc13. The structural basis for lysine 63 chain catalysis. J. Biol. Chem. 278, 13151–13158 (2003).
Miura, T., Klaus, W., Gsell, B., Miyamoto, C. & Senn, H. Characterization of the binding interface between ubiquitin and class I human ubiquitin-conjugating enzyme 2b by multidimensional heteronuclear NMR spectroscopy in solution. J. Mol. Biol. 290, 213–228 (1999).
Spyracopoulos, L., Lewis, M.J. & Saltibus, L.F. Main chain and side chain dynamics of the ubiquitin conjugating enzyme variant human Mms2 in the free and ubiquitin-bound states. Biochemistry 44, 8770–8781 (2005).
Lewis, M.J., Saltibus, L.F., Hau, D.D., Xiao, W. & Spyracopoulos, L. Structural basis for non-covalent interaction between ubiquitin and the ubiquitin conjugating enzyme variant human MMS2. J. Biomol. NMR 34, 89–100 (2006).
Vijay-Kumar, S., Bugg, C.E. & Cook, W.J. Structure of ubiquitin refined at 1.8 Å resolution. J. Mol. Biol. 194, 531–544 (1987).
Tsui, C., Raguraj, A. & Pickart, C.M. Ubiquitin binding site of the ubiquitin E2 variant (UEV) protein Mms2 is required for DNA damage tolerance in the yeast RAD6 pathway. J. Biol. Chem. 280, 19829–19835 (2005).
Piotrowski, J. et al. Inhibition of the 26 S proteasome by polyubiquitin chains synthesized to have defined lengths. J. Biol. Chem. 272, 23712–23721 (1997).
Wu, P.Y. et al. A conserved catalytic residue in the ubiquitin-conjugating enzyme family. EMBO J. 22, 5241–5250 (2003).
Reverter, D. & Lima, C.D. Insights into E3 ligase activity revealed by a SUMO-RanGAP1-Ubc9-Nup358 complex. Nature 435, 687–692 (2005).
Raasi, S. & Pickart, C.M. Rad23 ubiquitin-associated domains (UBA) inhibit 26 S proteasome-catalyzed proteolysis by sequestering lysine 48-linked polyubiquitin chains. J. Biol. Chem. 278, 8951–8959 (2003).
Haldeman, M.T., Xia, G., Kasperek, E.M. & Pickart, C.M. Structure and function of ubiquitin conjugating enzyme E2–25K: the tail is a core-dependent activity element. Biochemistry 36, 10526–10537 (1997).
Otwinowski, Z. & Minor, W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 176, 307–326 (1997).
Kissinger, C.R., Gehlhaar, D.K. & Fogel, D.B. Rapid automated molecular replacement by evolutionary search. Acta Crystallogr. D Biol. Crystallogr. 55, 484–491 (1999).
Jones, T.A., Zou, J.Y., Cowan, S.W. & Kjeldgaard, M. Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr A 47, 110–119 (1991).
Emsley, P. & Cowtan, K. Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132 (2004).
Brunger, A.T. et al. Crystallography & NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr. D Biol. Crystallogr. 54, 905–921 (1998).
Acknowledgements
We thank M. Bianchet, S. Gabelli and S. Bouyain for crystallographic assistance, B. Schulman, R. Cohen, B. Garcia-Moreno and C. Fitch for advice and discussions, and C. Ralston of Advanced Light Source beamline 8.2.2 for technical assistance. This work was supported by the Howard Hughes Medical Institute (C.W.) and US National Institutes of Health grant GM60372 (C.M.P.).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Fig. 1
SDS gel showing the ternary complex used for crystallization and the contents of the dissolved crystal. (PDF 71 kb)
Rights and permissions
About this article
Cite this article
Eddins, M., Carlile, C., Gomez, K. et al. Mms2–Ubc13 covalently bound to ubiquitin reveals the structural basis of linkage-specific polyubiquitin chain formation. Nat Struct Mol Biol 13, 915–920 (2006). https://doi.org/10.1038/nsmb1148
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nsmb1148
This article is cited by
-
cIAP1-based degraders induce degradation via branched ubiquitin architectures
Nature Chemical Biology (2023)
-
Ubiquitination in plant biotic and abiotic stress
Plant Growth Regulation (2023)
-
Ubiquitination in the rice blast fungus Magnaporthe oryzae: from development and pathogenicity to stress responses
Phytopathology Research (2022)
-
A new dawn beyond lysine ubiquitination
Nature Chemical Biology (2022)
-
Structural and functional asymmetry of RING trimerization controls priming and extension events in TRIM5α autoubiquitylation
Nature Communications (2022)