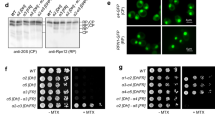
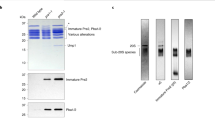
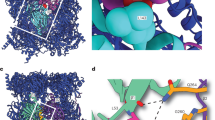
Abstract
Proteasome activity is fine-tuned by associating the proteolytic core particle (CP) with stimulatory and inhibitory complexes. Although several mammalian regulatory complexes are known, knowledge of yeast proteasome regulators is limited to the 19-subunit regulatory particle (RP), which confers ubiquitin-dependence on proteasomes. Here we describe an alternative proteasome activator from Saccharomyces cerevisiae, Blm10. Synthetic interactions between blm10Δ and other mutations that impair proteasome function show that Blm10 functions together with proteasomes in vivo. This large, internally repetitive protein is found predominantly within hybrid Blm10–CP–RP complexes, representing a distinct pool of mature proteasomes. EM studies show that Blm10 has a highly elongated, curved structure. The near-circular profile of Blm10 adapts it to the end of the CP cylinder, where it is properly positioned to activate the CP by opening the axial channel into its proteolytic chamber.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$189.00 per year
only $15.75 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout







Similar content being viewed by others
References
Pickart, C.M. & Cohen, R.E. Proteasomes and their kin: proteases in the machine age. Nat. Rev. Mol. Cell Biol. 5, 177–187 (2004).
Groll, M. et al. Structure of 20S proteasome from yeast at 2.4 Å resolution. Nature 386, 463–471 (1997).
Groll, M. et al. A gated channel into the proteasome core particle. Nat. Struct. Biol. 7, 1062–1067 (2000).
Braun, B.C. et al. The base of the proteasome regulatory particle exhibits chaperone-like activity. Nat. Cell Biol. 1, 221–226 (1999).
Liu, C.W. et al. Conformational remodeling of proteasomal substrates by PA700, the 19 S regulatory complex of the 26 S proteasome. J. Biol. Chem. 277, 26815–26820 (2002).
Schmidt, M., Lupas, A.N. & Finley, D. Structure and mechanism of ATP-dependent proteases. Curr. Opin. Chem. Biol. 3, 584–591 (1999).
Hartmann-Petersen, R., Tanaka, K. & Hendil, K.B. Quaternary structure of the ATPase complex of human 26S proteasomes determined by chemical cross-linking. Arch. Biochem. Biophys. 386, 89–94 (2001).
Prakash, S., Tian, L., Ratliff, K.S., Lehotzky, R.E. & Matouschek, A. An unstructured initiation site is required for efficient proteasome-mediated degradation. Nat. Struct. Mol. Biol. 11, 830–837 (2004).
Navon, A. & Goldberg, A.L. Proteins are unfolded on the surface of the ATPase ring before transport into the proteasome. Mol. Cell 8, 1339–1349 (2001).
Lam, Y.A., Lawson, T.G., Velayutham, M., Zweier, J.L. & Pickart, C.M. A proteasomal ATPase subunit recognizes the polyubiquitin degradation signal. Nature 416, 763–767 (2002).
Elsasser, S., Chandler-Militello, D., Mueller, B., Hanna, J. & Finley, D. Rad23 and Rpn10 serve as alternative ubiquitin receptors for the proteasome. J. Biol. Chem. 279, 26817–26822 (2004).
Verma, R., Oania, R., Graumann, J. & Deshaies, R.J. Multiubiquitin chain receptors define a layer of substrate selectivity in the ubiquitin-proteasome system. Cell 118, 99–110 (2004).
Yao, T. & Cohen, R.E. A cryptic protease couples deubiquitination and degradation by the proteasome. Nature 419, 403–407 (2002).
Leggett, D.S. et al. Multiple associated proteins regulate proteasome structure and function. Mol. Cell 10, 495–507 (2002).
Guterman, A. & Glickman, M.H. Complementary roles for Rpn11 and Ubp6 in deubiquitination and proteolysis by the proteasome. J. Biol. Chem. 279, 1729–1738 (2004).
Wojcik, C., Tanaka, K., Paweletz, N., Naab, U. & Wilk, S. Proteasome activator (PA28) subunits, α, β and γ (Ki antigen) in NT2 neuronal precursor cells and HeLa S3 cells. Eur. J. Cell Biol. 77, 151–160 (1998).
Ustrell, V., Hoffman, L., Pratt, G. & Rechsteiner, M. PA200, a nuclear proteasome activator involved in DNA repair. EMBO J. 21, 3516–3525 (2002).
Whitby, F.G. et al. Structural basis for the activation of 20S proteasomes by 11S regulators. Nature 408, 115–120 (2000).
Forster, A., Whitby, F.G. & Hill, C.P. The pore of activated 20S proteasomes has an ordered 7-fold symmetric conformation. EMBO J. 22, 4356–4364 (2003).
Tanahashi, N. et al. Hybrid proteasomes. Induction by interferon-γ and contribution to ATP-dependent proteolysis. J. Biol. Chem. 275, 14336–14345 (2000).
Doherty, K., Pramanik, A., Pride, L., Lukose, J. & Wood Moore, C. Expression of the expanded YFL007w ORF and assignment of the gene name BLM10. Yeast 21, 1021 (2004).
Fehlker, M., Wendler, P., Lehmann, A. & Enenkel, C. Blm3 is part of nascent proteasomes and is involved in a late stage of nuclear proteasome assembly. EMBO Rep. 4, 959–963 (2003).
Glickman, M.H., Rubin, D.M., Fried, V.A. & Finley, D. The regulatory particle of the Saccharomyces cerevisiae proteasome. Mol. Cell. Biol. 18, 3149–3162 (1998).
Kopp, F., Dahlmann, B. & Kuehn, L. Reconstitution of hybrid proteasomes from purified PA700-20 S complexes and PA28αβ activator: ultrastructure and peptidase activities. J. Mol. Biol. 313, 465–471 (2001).
Cascio, P., Call, M., Petre, B.M., Walz, T. & Goldberg, A.L. Properties of the hybrid form of the 26S proteasome containing both 19S and PA28 complexes. EMBO J. 21, 2636–2645 (2002).
Kajava, A.V., Gorbea, C., Ortega, J., Rechsteiner, M. & Steven, A.C. New HEAT-like repeat motifs in proteins regulating proteasome structure and function. J. Struct. Biol. 146, 425–430 (2004).
Groves, M.R., Hanlon, N., Turowski, P., Hemmings, B.A. & Barford, D. The structure of the protein phosphatase 2A PR65/A subunit reveals the conformation of its 15 tandemly repeated HEAT motifs. Cell 96, 99–110 (1999).
Chook, Y.M. & Blobel, G. Structure of the nuclear transport complex karyopherin-β2-Ran t · GppNHp. Nature 399, 230–237 (1999).
Vetter, I.R., Arndt, A., Kutay, U., Gorlich, D. & Wittinghofer, A. Structural view of the Ran-importin β interaction at 2.3 Å resolution. Cell 97, 635–646 (1999).
Goldenberg, S.J. et al. Structure of the Cand1-Cul1-Roc1 complex reveals regulatory mechanisms for the assembly of the multisubunit cullin-dependent ubiquitin ligases. Cell 119, 517–528 (2004).
Meiners, S. et al. Inhibition of proteasome activity induces concerted expression of proteasome genes and de novo formation of mammalian proteasomes. J. Biol. Chem. 278, 21517–21525 (2003).
London, M.K., Keck, B.I., Ramos, P.C. & Jurgen Dohmen, R. Regulatory mechanisms controlling biogenesis of ubiquitin and the proteasome. FEBS Lett. 567, 259–264 (2004).
Xie, Y. & Varshavsky, A. RPN4 is a ligand, substrate, and transcriptional regulator of the 26S proteasome: a negative feedback circuit. Proc. Natl. Acad. Sci. USA 98, 3056–3061 (2001).
Andrade, M.A., Petosa, C., O'Donoghue, S.I., Muller, C.W. & Bork, P. Comparison of ARM and HEAT protein repeats. J. Mol. Biol. 309, 1–18 (2001).
Moore, C.W. Further characterizations of bleomycin-sensitive (blm) mutants of Saccharomyces cerevisiae with implications for a radiomimetic model. J. Bacteriol. 173, 3605–3608 (1991).
Krogan, N.J. et al. Proteasome involvement in the repair of DNA double-strand breaks. Mol. Cell 16, 1027–1034 (2004).
Andrade, M.A., Perez-Iratxeta, C. & Ponting, C.P. Protein repeats: structures, functions, and evolution. J. Struct. Biol. 134, 117–131 (2001).
Kajava, A.V. Review: proteins with repeated sequence—structural prediction and modeling. J. Struct. Biol. 134, 132–144 (2001).
Chen, P. & Hochstrasser, M. Autocatalytic subunit processing couples active site formation in the 20S proteasome to completion of assembly. Cell 86, 961–972 (1996).
Forster, A. & Hill, C.P. Proteasome degradation: enter the substrate. Trends Cell Biol. 13, 550–553 (2003).
McCutchen-Maloney, S.L. et al. cDNA cloning, expression, and functional characterization of PI31, a proline-rich inhibitor of the proteasome. J. Biol. Chem. 275, 18557–18565 (2000).
Groettrup, M. et al. A role for the proteasome regulator PA28α in antigen presentation. Nature 381, 166–168 (1996).
Sijts, A. et al. The role of the proteasome activator PA28 in MHC class I antigen processing. Mol. Immunol. 39, 165–169 (2002).
Murata, S. et al. Growth retardation in mice lacking the proteasome activator PA28γ. J. Biol. Chem. 274, 38211–38215 (1999).
Brachmann, C.B. et al. Designer deletion strains derived from Saccharomyces cerevisiae S288C: a useful set of strains and plasmids for PCR-mediated gene disruption and other applications. Yeast 14, 115–132 (1998).
Goldstein, A.L. & McCusker, J.H. Three new dominant drug resistance cassettes for gene disruption in Saccharomyces cerevisiae. Yeast 15, 1541–1553 (1999).
Knop, M. et al. Epitope tagging of yeast genes using a PCR-based strategy: more tags and improved practical routines. Yeast 15, 963–972 (1999).
Longtine, M.S. et al. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast 14, 953–961 (1998).
Frank, J. et al. SPIDER and WEB: processing and visualization of images in 3D electron microscopy and related fields. J. Struct. Biol. 116, 190–199 (1996).
Ohi, M., Li, Y., Cheng, Y. & Walz, T. Negative staining and image classification—powerful tools in modern electron microscopy. Biol. Proceed. Online 6, 23–34 (2004).
Gerlinger, U.M., Guckel, R., Hoffmann, M., Wolf, D.H. & Hilt, W. Yeast cycloheximide-resistant crl mutants are proteasome mutants defective in protein degradation. Mol. Biol. Cell 8, 2487–2499 (1997).
Robben, J., Hertveldt, K., & Volckaert, G. Revisiting the yeast chromosome VI DNA sequence reveals a correction merging YFL007w and YFL006w to a single ORF. Yeast 19, 699–702 (2002).
Acknowledgements
This work was supported by a grant from the Deutsche Forschungsgemeinschaft to M.S. (SCHM 884/3-1), and US National Institutes of Health (NIH) grants to D.F. (GM65592) and S.G. (GM67945). We thank B. Petre for collecting the EM images. The molecular EM facility at Harvard Medical School was established by a generous donation from the Giovanni Armenise Harvard Center for Structural Biology and is maintained by funds from NIH grant GM62580 to T.W. We are also grateful to the Harvard Medical School Nikon Imaging Center for technical assistance and access to their instruments, to R. Li for providing us with an antibody against Arc15, and to S. Elsasser, J. Hanna and J. Roelofs for critical reading of the manuscript. We thank R. Gali, and the Bauer Center for Genomics Research, Harvard University, for help with preliminary results of microarray analysis on the Rosetta Resolver microarray analysis platform. We furthermore are grateful to C. Hill for communicating results prior to publication and to A.L. Goldberg for his permission to include PA28-20S electron micrographs in our study.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Fig. 1
Identification of CP-bound Blm10 by mass spectrometry. (PDF 20 kb)
Supplementary Fig. 2
Purification of CP from a blm10Δ strain. (PDF 316 kb)
Supplementary Fig. 3
Identification of RP-CP bound Blm10 by mass spectrometry. (PDF 39 kb)
Supplementary Fig. 4
Sequence alignment of the conserved Blm10 C terminus. (PDF 73 kb)
Supplementary Table 1
Primer used in this study. (PDF 12 kb)
Rights and permissions
About this article
Cite this article
Schmidt, M., Haas, W., Crosas, B. et al. The HEAT repeat protein Blm10 regulates the yeast proteasome by capping the core particle. Nat Struct Mol Biol 12, 294–303 (2005). https://doi.org/10.1038/nsmb914
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nsmb914
This article is cited by
-
The ubiquitin–proteasome system in kidney physiology and disease
Nature Reviews Nephrology (2019)
-
Proteasome activators, PA28γ and PA200, play indispensable roles in male fertility
Scientific Reports (2016)
-
Phosphorylation of the C-terminal tail of proteasome subunit α7 is required for binding of the proteasome quality control factor Ecm29
Scientific Reports (2016)
-
Maturation of the proteasome core particle induces an affinity switch that controls regulatory particle association
Nature Communications (2015)
-
MiR-29b replacement inhibits proteasomes and disrupts aggresome+autophagosome formation to enhance the antimyeloma benefit of bortezomib
Leukemia (2015)