Abstract
Neurokinins (or tachykinins) are peptides that modulate a wide variety of human physiology through the neurokinin G protein-coupled receptor family, implicated in a diverse array of pathological processes. Here we report high-resolution crystal structures of the human NK1 receptor (NK1R) bound to two small-molecule antagonist therapeutics – aprepitant and netupitant and the progenitor antagonist CP-99,994. The structures reveal the detailed interactions between clinically approved antagonists and NK1R, which induce a distinct receptor conformation resulting in an interhelical hydrogen-bond network that cross-links the extracellular ends of helices V and VI. Furthermore, the high-resolution details of NK1R bound to netupitant establish a structural rationale for the lack of basal activity in NK1R. Taken together, these co-structures provide a comprehensive structural basis of NK1R antagonism and will facilitate the design of new therapeutics targeting the neurokinin receptor family.
Similar content being viewed by others
Introduction
The neurokinin (NK) receptor-ligand system represents a complex, evolutionarily conserved neuropeptide signaling architecture1,2. Derived from alternate processing of two genes, the best-characterised mammalian neurokinins are Substance P (SP), NKA and NKB, sharing the conserved C-terminal structural motif FxGLM-NH23. Among other neurokinins, these three peptides act as agonists with different affinities and selectivity for three pharmacologically distinct neurokinin receptors4 (NK1R, NK2R and NK3R) that belong to the superfamily of G protein-coupled receptors (GPCRs). Within this system, SP represents the preferred endogenous agonist of NK1R4. NK1R has been shown to be present in the central and peripheral nervous system5,6, smooth muscle7, endothelial cells8 and also on cells that participate in the immune response9. Over the past four decades, intensive research has linked the SP-NK1R system to such diverse pathophysiological processes as nausea10, analgesia11,12, inflammation13, pruritus14 and depression15,16, highlighting the potential therapeutic value of antagonists directed against NK1R. This prospect triggered widespread efforts across industry and academia to discover such compounds to date17,18.
Disclosure of the first non-peptide NK1R antagonist CP-96,34519 (Supplementary Figure 1), discovered by high-throughput screening (HTS), subsequently spurred the development of a number of antagonists with improved pharmacological properties. This led to the identification of CP-99,99420, which reduced the chemical structure of CP-96,345 to a molecular scaffold found in many later-stage small-molecule antagonists. CP-99,994 displays high affinity and selectivity for NK1R, as well as efficacy in animal models, and therefore historically provided a valuable pharmacological tool for the investigation of the physiological role of SP-mediated signaling through NK1R21. Modification of the central saturated six-membered piperidine ring of CP-99,994 alongside further scaffold optimisations22,23 (by substituent addition and modifications) ultimately lead to the development of aprepitant24 (Supplementary Figure 1), which became the first approved oral drug to make it into the clinic, specifically targeting NK1R for the treatment of chemotherapy-induced nausea and vomiting (CINV)25. Over the past four years, two further molecules (i.e. netupitant and rolapitant) that do not share the common chemical scaffold of these earlier antagonists have been approved for use in the clinic for the same indication17.
Extensive structure-activity relationship (SAR) studies performed over the course of three decades have revealed insights into overlapping and non-overlapping binding sites involved in recognition of peptide agonists and non-peptide antagonists in NK1R26,27,28. However, until now, little has been known about the precise binding mode of small-molecule antagonists to NK1R in the absence of a structure of this receptor. Since the prototypical antagonist CP-99,994 already displays many chemical features that are relatively preserved in further developed compounds we initially solved the structure of NK1R in complex with this ligand. We then went on to co-crystallise NK1R with two FDA-approved drugs—aprepitant and netupitant.
Here, we report three crystal structures of the human NK1R bound to CP-99,994 and the clinically approved antagonists aprepitant and netupitant at 3.27, 2.40 and 2.20 Å resolution, respectively. These structures provide detailed and high-resolution structural insights into the molecular determinants of NK1R antagonist recognition. The clinically approved antagonists are able to invoke structural rearrangements in the orthosteric binding pocket at the extracellular ends of helices V and VI and the extracellular loops (specifically ECL2) that govern the overall size and nature of the pocket, thereby acting to negatively modulate the receptor via an induced-fit binding mechanism. The observed high degree of plasticity in the NK1R orthosteric binding pocket across the three structures reported here vastly improves our structural knowledge of NK1R, explaining the different properties of current inhibitors and potentially facilitating the future development of ligands selectively targeting various NK receptors.
Results
Crystallisation of antagonist-bound NK1R
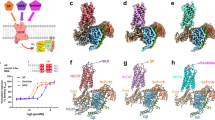
To improve protein expression, and ultimately the yield of NK1R preparations, two consecutive rounds of directed evolution in Saccharomyces cerevisiae were initially performed on the human NK1R29. One evolved receptor mutant (NK1R-y04) was further thermostabilised in an antagonist-bound state through incorporation of four amino acid substitutions, L742.46A, A1444.39L, A2155.57L and K2436.30A (Ballesteros and Weinstein numbering30 denoted in superscript), leading to NK1RS (Methods and Supplementary Figure 2a, b). To facilitate crystallisation in lipidic cubic phase, 11 residues (E227-H237) of the third intracellular loop (ICL3) were replaced by the thermostable PGS (Pyrococcus abysii glycogen synthase) domain31. The crystallised PGS fusion construct NK1RXTAL is able to bind all co-crystallised antagonists with low nanomolar affinity. However, ligand affinity is reduced ∼10-fold when compared to the wild-type receptor, possibly due to an increased rigidity of the stabilised fusion construct (Supplementary Table 1). For crystallisation in lipidic cubic phase, NK1RXTAL was purified from Spodoptera frugiperda (Sf9) insect cell membranes in the presence of either CP-99,994, aprepitant or netupitant. We then crystallised and determined three crystal structures of NK1R bound to three different antagonists, complexed with CP-99,994 at 3.27 Å resolution, aprepitant at 2.40 Å resolution and finally with netupitant at 2.20 Å resolution (Table 1), with strong and unambiguous electron density for each antagonist present in the orthosteric site as well as key interaction residues of the receptor (Fig. 1d–f and Supplementary Figure 3d–i).
Overall structures of antagonist-bound NK1R. a–c NK1R in complex with CP-99,994 (a), aprepitant (b) and netupitant (c), viewed parallel to the membrane plane. The receptors are depicted by ribbons and coloured in brown, turquois and blue, respectively. The ligands CP-99,994, aprepitant and netupitant are shown as spheres and coloured in pink, yellow and orange, respectively. Oxygen, nitrogen and fluorine atoms of the ligands are highlighted in red, blue and grey, respectively. d–f NK1R in complex with CP-99,994 (d), aprepitant (e) and netupitant (f), viewed from the extracellular space and coloured as in (a–c). The ligands are depicted as sticks. 2Fo-Fc electron density maps of the ligands are shown in green mesh contoured at 1.0 σ
While NK1R in complex with CP-99,994 crystallised in space group C2221 (Supplementary Figure 4b, d–f), it is noteworthy that crystallisation of NK1R with both aprepitant and netupitant consistently lead to better diffracting crystals in a different condition belonging to space group P212121 (Supplementary Figure 4c, g–i). However, attempts to switch crystallisation conditions, i.e. crystallisation of NK1R with CP-99,994 in those specific to netupitant or aprepitant, were not successful, indicating that the receptor conformations described here are specific to the ligands with which they are crystallised.
Overall architecture of NK1R
Overall, NK1R exhibits the canonical GPCR architecture comprising seven transmembrane helices (I–VII) with helix 8 lying parallel to the membrane plane (Fig. 1a–c). All intracellular and extracellular loops (ICLs and ECLs, respectively) are well resolved with the exception of ECL3. In NK1R, similar to other structures of the β-branch of class A GPCRs, ECL2 forms an extended β-hairpin crossing above the orthosteric pocket and is anchored to the extracellular tip of helix III through a conserved disulfide bridge between C180ECL2 and C1053.25.
The structure of the transmembrane helical bundle of NK1R is similar to those of other receptors from the β-branch of class A GPCRs bound to small-molecule antagonists, with root-mean-square deviations (RMSD) for backbone atoms of 1.3 Å to orexin 2 receptor31 (OX2R) (PDB ID 4S0V), 1.4 Å to neuropeptide Y Y1 receptor32 (Y1R) (PDB ID 5ZBH) and 2.5 Å to endothelin B receptor33 (ETBR) (PDB ID 5XPR) (Supplementary Figure 5a–i). In the three NK1R structures, the highly conserved residue W2616.48, which has been reported as the “toggle switch” important in triggering GPCR activation34, is in a similar conformation to those observed in other inactive class A GPCR structures. Furthermore, the “ionic lock” involving R3.50 of the D/ERY motif, making an intrahelical salt bridge with D3.49, is present in all solved NK1R structures. Taken together, the NK1R structures in complex with different antagonists capture the inactive receptor conformation.
The NK1R orthosteric antagonist binding pocket
In all three NK1R co-structures, the antagonist small molecules are found buried within a largely hydrophobic cleft constituting the orthosteric binding pocket. The binding site is characterised by being elongated on a trajectory between helices II and IV and laterally constricted by side chains of helices III and VI (Fig. 1d–f and Supplementary Figure 6a–c). Although the overall shape of the NK1R orthosteric binding pocket is itself unique, its depth and overall location within the transmembrane helical bundle is comparable to those in previously reported closely related structures, confirming that the non-peptide NK1R small molecule antagonists occupy at least in part the typical drug-binding site of peptide-activated receptors (Supplementary Figure 5a–i).
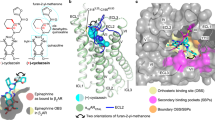
Most high-affinity NK1R-selective antagonists are characterised by a common pharmacophore. In general, this structural framework consists of a central six-membered ring with two (and in later-stage compounds three) bulky substituents (Fig. 2d–f). Henceforth, and for clarity, the central ring is termed the “core”, the linker-attached bulky aromatic group (i.e. the methoxyphenyl group in CP-99,994 or di-trifluoromethylphenyl in all other antagonists) “arm 1” and the directly attached aromatic moiety (i.e. the phenyl in CP-99,994 or fluorophenyl in aprepitant or methylphenyl in netupitant) “arm 2”. The additional variable cyclic substituent at the core present in aprepitant and netupitant is termed “arm 3”. Since CP-99,994 represents the progenitor antagonist on which many of the later developed compounds were based (and as such makes only limited contact to the receptor) the interactions of CP-99,994 within the NK1R binding site are described first.
Drug-binding site of NK1R. a–c Detailed interactions of CP-99,994 (a), aprepitant (b) and netupitant (c) with the receptor, viewed from the extracellular side from a position above helix I. The receptor backbone is shown in grey ribbon representation. Ligand and receptor residues within 4 Å of the respective antagonist are shown as sticks and are coloured as in Fig. 1. The ordered water involved in netupitant binding is depicted as a red sphere (c). Hydrogen bonds are indicated by dashed blue lines. d–f Chemical structures of CP-99,994 (d), aprepitant (e) and netupitant (f) with structural topology highlighted by coloured, dashed circles (core coloured in blue, arm 1 in yellow, arm 2 in green, arm 3 in red, respectively). g–i Antagonist affinity profiles of selected mutants in comparison to wild-type NK1R. pKi values for each antagonist were derived from competition ligand-binding experiments (Supplementary Table 1). Bars represent differences in calculated affinity (pKi) values for each mutant relative to the wild-type receptor for CP-99,994 (g), aprepitant (h), and netupitant (i). Data are shown as mean values ± s.e.m. from three to five independent experiments performed in duplicates. n.b. no binding. Source Data
CP-99,994 specifically interacts with only seven residues from helices III to VI and one residue from ECL2 in NK1R (Fig. 2a and Supplementary Figure 3a). The core (2,3-cis-substituted-piperidine) is laterally wedged between the side chains of F2686.55 and Q1654.60 and capped by I182ECL2. Q1654.60 is positioned to concomitantly hydrogen-bond the piperidine core and the amine linker of arm 1. Mutation of Q1654.60 to either alanine, glutamic acid or aspartic acid severely reduce the binding affinity of CP-99,994 (Fig. 2g). These data are in agreement with previous studies reporting the critical nature of this interaction in binding of CP-99,994 and other early non-peptide antagonists to NK1R35. Mutation of F2686.55 to alanine attenuates binding affinity more than 10-fold36, highlighting the crucial nature of the hydrophobic stacking interactions with the small-molecule core moiety on this side of the orthosteric pocket.
Arm 1 of CP-99,994 extends deep into the receptor core, exploiting a large, almost exclusively lipophilic groove at the base of the orthosteric pocket formed by M2917.39, M2957.43, M812.53, W2616.48, I2045.46, F2646.51 and P1123.32. Within this region, the aromatic methoxyphenyl group of arm 1 is wedged between F2646.51 and P1123.32 and sits above a deep protrusion into the receptor core. The methoxy group points into a relatively spacious extension of this pocket comprised of residues from helices II, III and VII. Mutation of P1123.32 to either aspartic acid or histidine results in a 4000-fold loss in binding affinity of CP-99,994 to NK1R36 with mutation of F2646.51 to alanine displaying only a moderate (4-fold) impairment of binding35, highlighting the importance of targeting this lipophilic local environment. However, the importance of this sub-pocket is underlined by the introduction of a more bulky tryptophan at this position leading to a >15-fold reduction in binding affinity for CP-99,994 (Fig. 2g).
Arm 2 of CP-99,994 targets a second lipophilic sub-pocket, making hydrophobic contacts to I1133.33 and V2005.42, and is capped on top by a face-on π-stack with H1975.39. Mutation of H1975.39 to alanine attenuates binding affinity 3.5-fold, yet preserving the aromatic character of this residue by virtue of mutation to phenylalanine results in only a <2-fold reduction in binding affinity to the small molecule, supporting this observed π-π interaction with the receptor.
Aprepitant, with its more extended structure due to its arm 3 substituent, engages in a host of additional interactions with NK1R (Fig. 2b and Supplementary Figure 3b). The core of aprepitant (2,3,4-substituted-morpholine) is situated between F2686.55 and Q1654.60 and capped by a hydrophobic interaction with I182ECL2 as in CP-99,994. However, Q1654.60 does not contact the morpholine core, instead making hydrogen-bonds to the oxygen atom of the arm 1 ether linker and a nitrogen atom of the arm 3 substituent. The methyl substituent of the arm 1 ether linker then optimally targets a small lipophilic subpocket formed between N1093.29, P1123.32 and I1133.33 on the surface of helix III.
The aromatic ring of arm 1 is shifted upwards and sideways towards helix II in comparison to CP-99,994. This arrangement allows it to optimally exploit the hydrophobic base of this part of the orthosteric pocket immediately above and adjacent to W2616.48 by virtue of the 3,5-di-trifluoromethyl groups that straddle the aromatic side chain of this residue. It is possible that this direct engagement acts to prevent the activation-related motion of W2616.48, thereby further stabilising NK1R in the inactive conformation.
In a similar fashion to CP-99,994, arm 2 of aprepitant targets a lipophilic subpocket formed by residues H1975.39, V2005.42, T2015.43, I2045.46 and H2656.52. However, the additional arm 3 triazolinone substituent of aprepitant is found creating an extended binding pocket (EBP) between the extracellular ends of helices IV, V and VI and ECL2. Aprepitant can thus engage an array of additional interactions with NK1R. The arm 2 (fluorophenyl) and arm 3 (triazolinone) substituents of aprepitant participate in a π-stacking interaction with H1975.39, while the carbonyl group of the triazolinone group reaches up to hydrogen-bond with the indole nitrogen of W184ECL2, with E1935.35 hydrogen-bonding to the N3 atom of the triazolinone ring. On the other side of the triazolinone ring, the carbonyl group of Q1654.60 makes a hydrogen bond with N5 of this 5-membered ring. As a likely consequence of the additional network of interactions that aprepitant makes to the receptor, the affinity of this small molecule is much less affected by single point mutations in comparison to CP-99,994 (Fig. 2h). Mutation of Q1654.60 to alanine, glutamic acid or aspartic acid only maximally impairs binding <5-fold, in agreement with earlier findings in which the mutant Q1654.60A displayed a 10-fold decrease in binding affinity for the close aprepitant analogue L-742,69423. In contrast to CP-99,994, aprepitant therefore induces a conformational change of ECL2 and the extracellular ends of helix V and VI, highlighting an unexpected high degree of plasticity across this part of the receptor.
Both NK1R antagonists described thus far are based on the chiral pharmacophore core of CP-99,994. In contrast, netupitant, which was disclosed in 2006 and clinically approved in 201437, belongs to a class of achiral, high-affinity NK1R antagonists with an aromatic pyridine core38. In a similar fashion to aprepitant, netupitant adopts an elongated conformation inside the orthosteric binding site (Fig. 2c); however, it protrudes from the centre of the orthosteric binding site towards extracellular space with both its core and arm 3 substituent. The di-trifluoromethylphenyl group of arm 1 and the methylphenyl group of arm 2 both target the same hydrophobic pockets as the equivalent substituents of the chiral antagonists. However, arm 2 is found to be in a more “upright” position. Major deviations in the positioning of the core region are observed within the orthosteric binding site, the linker region of arm 1, and the arm 3 substituent.
Due to the elongated linker of arm 1, the aromatic core of netupitant is pushed upwards and towards extracellular space from the centre of the pocket (towards helices V and VI) (Fig. 2c and Supplementary Figure 6c), while also moving sideways into closer proximity of F2686.55. This observed repositioning of the substituted pyridine towards helix V and in particular into closer proximity of H1975.39 leads to an edge-to-face π-π interaction between this side chain and the aromatic core. Consequently, the importance of this aromatic interaction is reflected in a 10-fold loss in netupitant binding affinity upon mutation of H1975.39 to alanine. In contrast, preserving the aromatic nature at this position with a H1975.39F mutation leaves the binding affinity unchanged (Fig. 2i and Supplementary Table 1). In the netupitant-bound NK1R structure, the side chain of Q1654.60 is rotated by 180° (compared to all other structures), enabling the water-mediated coordination of the oxygen atom of the amide located in the arm 1 linker (Fig. 2c and Supplementary Figure 3c). Furthermore, due to the different ligand position within the binding site, arm 3 of netupitant targets a small hydrophobic groove formed between residues F2686.55, P2716.58 and Y2726.59 at the extracellular tip of helix VI. The opening of this cleft between two helical turns is supported by a slight outward bending of the extracellular tip of helix VI towards the lipid bilayer. Overall, with the exception of the direct π-π interaction between the core and H1975.39, the additional interactions between netupitant and NK1R render the affinity of the antagonist less affected by single point mutations when compared to their influence on CP-99,994 binding affinity (Fig. 2i).
Conformational changes induced by clinical NK1R antagonists
The overall structure of netupitant-bound NK1R is similar to that of the aprepitant-bound structure with an all-atom RMSD of ∼0.40 Å (Supplementary Figure 7a). However, both the aprepitant- and netupitant-bound NK1R structures differ from the CP-99,994-bound structure with all-atom RMSDs of ∼0.70 Å. In comparison to the CP-99,994-bound receptor structure, aprepitant induces structural rearrangements in the extracellular regions of NK1R, and thus modulates the extracellular molecular landscape of the receptor (Fig. 3a and Supplementary Figure 7b): The β-sheet of ECL2 is kinked away from the extracellular end of helix III by 4.1 Å (as measured between equivalent Cα atoms of T173ECL2), thereby accommodating an inward movement of helix II and ECL1 by 1.7 Å (as measured between equivalent Cα atoms of H952.67) and 2.1 Å (as measured between equivalent Cα atoms of E97ECL1), respectively. In addition, the β-hairpin loop is itself twisted towards the central axis of the receptor. This appears to be related to the observed cis-configuration of the peptide bond preceding P175ECL2 located at the turn of the β-hairpin (Supplementary Figure 7c–e), while a trans-proline is found in the CP-99,994-bound structure. Together, these differences lead to a slight contraction of the orthosteric binding pocket at the helix II–IV interface of the receptor when binding aprepitant or netupitant.
Conformational changes in NK1R induced by clinically used antagonists. a Superimposition of the CP-99,994- and aprepitant-bound NK1R structures, viewed from helix I. Residues of NK1R with side-chain orientations differing between the two receptor structures as well as the antagonists are depicted as sticks, coloured as in Fig. 1. b, c Hydrogen bond network connecting the extracellular ends of helices V and VI in the aprepitant- (b) and netupitant-bound (c) NK1R structures as viewed from the membrane plane. Hydrogen bonds are indicated as dashed blue lines. d–f Close-up views on residues with differing side-chain orientation in the CP-99,994- and the aprepitant-bound NK1R structure. Side-chain rearrangements from the CP-99,994- to the aprepitant-bound conformation are indicated by black arrows
On the opposite side of the NK1R orthosteric hemisphere, the extracellular end of helix V together with the C-terminal portion of ECL2 is shifted 2.4 Å away from the central axis of the receptor (as measured between equivalent Cα atoms of K190ECL2). This shift is induced by the arm 3 substituent of aprepitant, which creates an EBP at the interface between helices IV, V and ECL2, thereby inducing significant side-chain rearrangements on this side of the receptor (Fig. 3a). The outward movement of the C-terminal part of ECL2 starts at W184ECL2, which is pushed 3.5 Å out of the interhelical interface between helices IV and V by the triazolinone group of aprepitant (Fig. 3f) in comparison to the CP-99,994-bound structure. The outward push of W184ECL2 is accommodated by a 90° rotation of Y1925.34 towards the lipid bilayer.
Further rearrangements caused by aprepitant are observed at the extracellular portion of helices V and VI. Firstly, K1945.36 is moved ~90° out of the orthosteric pocket, disengaging from E1935.35 which is then free to hydrogen-bond the triazolinone ring of aprepitant (Fig. 3e). Secondly, the side chain of H1975.39 is rotated by ~90° out of the orthosteric pocket, while Y2726.59 at the top of helix VI is moved by ~90° into the interface between helices V and VI (Fig. 3d). This rearrangement of H1975.39 and Y2726.59 causes the formation of an extended interhelical hydrogen bond network connecting residues Y2726.59, H1975.39, T2015.43 and H2656.52, thereby cross-linking the extracellular ends of helices V and VI (Fig. 3b).
Although the arm 3 substituent of netupitant targets a part of the receptor that is distinct from aprepitant (see above), both antagonists are found to induce the same hydrogen bond network by virtue of the reorientation of H1975.39 and Y2726.59 (Fig. 3b, c). However, in contrast to aprepitant, in the case of netupitant, the reorientation of H1975.39 is not caused solely by arm 3, but rather by the different binding pose of the compound within the orthosteric pocket, which shifts both the aromatic core and arm 2 into a position closer to helix V. This shift sterically precludes any other rotameric orientation of H1975.39 except the one observed in the crystal structure found engaged with the extended hydrogen bond network.
Interestingly, H1975.39 has previously been linked to play a role in insurmountable antagonism23. In that study, a single point mutation of H1975.39 to serine was shown to alter the mode of antagonism of a close aprepitant analogue (L-742,694, Supplementary Figure 1) from insurmountable to surmountable while only moderately affecting binding affinity. Furthermore, it was demonstrated that tethering of the tips on helix V and VI via engineering of a high-affinity metal ion-binding site involving H1975.39 and two histidine residues introduced at position E1935.35 and Y2726.59 rendered the receptor in an inactive conformation39. Finally, this region of helix V, and rearrangements of the hydrogen-bonding network between conserved serines on helix V (specifically residues 5.42, 5.43 and 5.46 which are a valine, threonine and isoleucine in NK1R, respectively) by the catechol group of epinephrine in β1-AR and β2-AR further points to the importance of this region on the extracellular side of helix V in controlling the constellation of GPCR functional states40,41.
It has long been acknowledged that interhelical hydrogen bonds have a strong influence on the conformational stability of membrane proteins42,43,44. The newly created hydrogen bond network, in the aprepitant-bound and netupitant-bound structures, tethers helix V to helix VI (Fig. 3b, c). In particular certain residues within the network have been shown to be critically involved in GPCR receptor activation41,45,46. We thus hypothesise that the reduction in conformational flexibility through engagement of the “histidine-lock” might represent a key driver for the observed insurmountable antagonism that some compounds such as aprepitant and netupitant elicit at NK1R.
Once the “histidine-lock” is engaged by structural rearrangements induced by the mentioned insurmountable antagonists, the receptor would be present in the proposed slow reversible state47,48, while in the absence of tethering helices V and VI with a hydrogen bond network only the fast reversible state would be populated (as observed in the CP-99,994-bound NK1R structure). Moreover, this structural differentiation might also be the basis of the prolonged in vivo efficacy of compounds such as aprepitant and netupitant49,50: once the lock is engaged, the distinct conformational state of the receptor may facilitate rebinding of compounds without the need for conformational rearrangements, thereby potentially increasing the effective association rate.
Structural basis of tight signaling control at NK1R
In the available high-resolution structures of inactive class A GPCRs where water molecules can be resolved, a conserved water-mediated hydrogen bond network is found to connect helices II, III, VI and VII51,52 (Supplementary Figure 8b–e). This network is often clustered around a central sodium ion coordinated between the highly conserved D2.50 and a polar residue on helix III (X3.39); concomitantly, sodium has been extensively described as a negative allosteric modulator stabilising the inactive receptor conformation of class A GPCRs53,54.
D2.50 is highly conserved (98%) across all class A GPCRs, with NK1R being one of the very few exceptions where an aspartic acid is not present at this position55. In NK1R, this position is occupied by a glutamate residue, E782.50 (Fig. 4a, b). Recent functional studies56 suggest that this sequence variant is linked to the unusual lack of constitutive signaling in NK1R27,36,57. The high resolution of the netupitant-bound NK1R structure reported here now provides a structural rationale for this observation. E782.50 occupies a more central position in the water-mediated hydrogen bond network compared to D2.50, making direct hydrogen bonds to S1193.39 and N3017.49 of helices III and VII, respectively (Fig. 4b). This stands in contrast to previously reported GPCR structures, where the contact between the D2.50 carboxyl group and the side chain of X3.39 is mediated by the aforementioned sodium ion58,59,60,61 (Supplementary Figure 8b–f) or potentially a water molecule33. Importantly, these features occur in addition to other conserved components of the network; for example, there is still a water-mediated hydrogen bond between the E782.50 side chain and the N3017.49 backbone nitrogen, as well as another between N3017.49 and V2536.40, recapitulating the interactions observed in the high-resolution D4R antagonist structure58 (Fig. 4c). Finally, a water-mediated hydrogen bond exists between helices I and II, through the side-chain oxygen atom of N501.50 and the carboxyl group of E782.50, similar to the interaction observed in several structures of other GPCRs such as the δ-OR59, β1-AR60 and A2AR61 (Supplementary Figure 8c–e).
E782.50-mediated interhelical hydrogen-bond network in the transmembrane core. a Amino acid sequence alignment of residues involved in the allosteric sodium ion-binding site in NK1R and other class A GPCRs (Ballesteros-Weinstein numbers indicated). b, c Extracellular view on the hydrogen-bonding network in the transmembrane core of netupitant-bound NK1R (b) and the dopamine D4 receptor (D4R, PDB ID 5WIV) (c). The receptor backbone is shown in ribbon representations coloured blue and yellow, respectively. Ordered waters are shown as red spheres and hydrogen bonds are indicated by dashed blue lines. The sodium ion coordinated between D2.50 and S3.39 in the D4R is shown as a purple sphere
Therefore, it appears that the direct interactions mediated by the larger glutamate side chain of NK1R at position X2.50 link the helical bundle of NK1R together more tightly than an aspartic acid would at the equivalent position, thus contributing to the stability of the inactive state of NK1R. Importantly, a recent study has shown that mutating E782.50 to aspartic acid results in a NK1 receptor phenotype which is susceptible to a similar sodium concentration-dependent negative allosteric effect as many other class A GPCRs56. Thus, E782.50 appears to almost assume the functional role of the sodium ion in other GPCRs by virtue of one single extra carbon atom—and in fact, may further enhance, strengthen and expand the key sodium-mediated interactions in providing a structural basis for the observed lack of basal signaling in NK1R56.
Discussion
The crystal structures of NK1R with CP-99,994 (the progenitor compound) and with aprepitant and netupitant, in use in the clinic today for the treatment of CINV, reveal the structural basis of the successful high-throughput screening and medicinal chemistry campaigns directed towards this receptor spanning several decades. To recapitulate, modifications of arm 1 and arm 2 substituents lead to a more optimal interaction with the narrow orthosteric binding pocket, while the additional arm 3 substituent present in aprepitant and netupitant allows these compounds to induce a distinct receptor conformation by a potential induced-fit binding mechanism.
Additionally, the structures provide a possible molecular rationale for the observed insurmountable antagonism of aprepitant and netupitant. These compounds create an inter-helical hydrogen-bonding network at the extracellular ends of helices V and VI, engaging a “histidine lock” in NK1R across a region known a priori to be important in modulating activity for other class A GPCRs. Finally, a mechanistic basis for the lack of basal signaling of NK1R is provided by the network of hydrogen bonds mediated by the unique residue E782.50 in the helical core of the receptor, which overlaps the allosteric sodium binding site observed in many inactive class A GPCR structures to date.
In light of the research presented here, it is now possible to deploy structure-based drug-design methods for designing new small molecules directed to NK1R, for the aforementioned indications. Furthermore, while the residues in the lower half of the orthosteric pocket are largely conserved between NK1R, NK2R and NK3R, sequence homology weakens towards the top of helix V and VI between these closely related receptors. With careful molecular modelling, coupled to molecular dynamics simulations, the structure of NK1R now opens new avenues to design the next generation of selective antagonists for the closely related NK2R and NK3R, by specifically targeting this region at the top of helices V and VI. The potential indications range from central neurological disorders including major depressive disorder and schizophrenia62,63, to vasomotor symptoms associated with the menopause64. Additionally, endosome-targeted NK1R small-molecule antagonists have recently been reported as pain relief agents, presenting an alternative to opioid-based drugs18. Since endosomal targeting as of now requires the coupling of the antagonist to a lipophilic moiety, our results will aid the rational design of new drugs in this class. In summary, the reported NK1R structures greatly contribute to our knowledge and understanding of how this clinically relevant family of receptors may be antagonised, providing opportunities to improve in vivo efficacy and reduce phase II attrition rates in the clinic for small-molecules directed towards these receptors.
Methods
Generation of NK1R crystallisation construct
From a pool of previously published NK1R mutants obtained by directed evolution in yeast for improved expression levels29 using HiLyte Fluor 488-labeled Substance P, NK1R-y04 (V1163.36I, M181ECL2K, W2245.66R, ΔC336-407) was selected as a potential crystallisation candidate since it could be well purified from Spodoptera frugiperda (Sf9) insect cells as a monodisperse protein. However, NK1R-y04 displayed only limited thermal stability in the CPM assay65 in complex with CP-99,994. To increase the thermostability of NK1R-y04 in an antagonist-bound state, selected amino acids within the transmembrane helical bundle were substituted with either alanine or leucine (if the amino acid was an alanine). The resulting single mutants were evaluated based on their gain in thermostability as evidenced by an increase in melting temperature (Tm) in the CPM assay when purified in the presence of CP-99,994. Single point-mutations which displayed the highest apparent gain in Tm (L742.46A, A1444.39L, A2155.57L and K2436.30A) were subsequently combined yielding the thermostabilised quadruple mutant NK1RS. Furthermore, to aid crystallisation in lipidic cubic phase, eleven residues (E227-H237) of the ICL3 were replaced by the thermostable PGS (Pyrococcus abysii glycogen synthase) domain, yielding NK1RXTAL. The final construct was cloned into a modified pFL vector (MultiBac system, Geneva Biotech) resulting in an expression construct with a melittin signal sequence followed by a FLAG-tag, His10-tag and a human rhinovirus 3C protease cleavage site N-terminal to the receptor gene (all primers used in this study are listed in Supplementary Table 2).
Expression and purification of NK1R
Recombinant baculovirus was generated using the MultiBac expression system. The receptor expression cassettes were integrated into the DH10EMBacY baculovirus genome and the resulting bacmids were transfected into Sf9 cells in 6-well tissue culture plates (2 ml, density of 4 × 105 cells/ml) using 8 μl of Cellfectin II Reagent (Thermo Fisher Scientific) and Sf-900 II SFM medium (Thermo Fisher Scientific). Viral P0 stocks were harvested as the supernatant after 4 days and were subsequently amplified to obtain high-titer viral P1 stocks. For expression, Sf9 insect cells in Sf-900 II SFM medium were infected with P1 virus at a cell density of 3 × 106 cells/ml and a multiplicity of infection of 5. Expression was performed for 72 h at 27 °C under constant shaking. Cells were harvested by centrifugation, washed with PBS, frozen in liquid nitrogen and stored at −80 °C.
Insect cells expressing NK1RXTAL were lysed and receptor-containing membranes isolated by repeated Dounce homogenisation in hypotonic (10 mM HEPES pH 7.5, 20 mM KCl, 10 mM MgCl2, 50 µg/ml Pefabloc SC (Carl Roth), 1 µg/ml Pepstatin A(Carl Roth)) and hypertonic buffer (10 mM HEPES pH 7.5, 20 mM KCl, 10 mM MgCl2, 1.0 M NaCl, 50 µg/ml Pefabloc SC, 1 µg/ml Pepstatin A). Purified membranes were resuspended in 30 ml hypotonic buffer supplemented with 40 µM of the respective antagonist (CP-99,994 (Tocris)/aprepitant (Sigma Aldrich)/netupitant (Selleckchem)), frozen in liquid nitrogen and stored at −80 °C until further use.
Frozen membranes were thawed on ice, the respective ligand (CP-99,994, aprepitant or netupitant) was added to a final concentration of 80 µM and the suspension was incubated for 30 min while turning. 2 mg/ml iodoacetamide (Sigma Aldrich) was added to the solution followed by another 30 min of incubation. Subsequently, the receptor was solubilised in 30 mM HEPES pH 7.5, 500 mM NaCl, 10 mM KCl, 5 mM MgCl2, 50 µg/ml Pefabloc SC, 1 µg/ml Pepstatin A, 1% (w/v) n-dodecyl-β-D-maltopyranoside (DDM, Anatrace) and 0.2% (w/v) cholesteryl hemisuccinate (CHS, Sigma Aldrich) at 4 °C for 3 h. Insoluble material was removed by ultra-centrifugation and the supernatant was incubated with TALON IMAC resin (GE Healthcare) at 4 °C overnight.
The receptor-bound resin was washed with 30 column volumes (CV) of Wash Buffer I (50 mM HEPES pH 7.5, 500 mM NaCl, 10 mM MgCl2, 5 mM imidazole, 10% (v/v) glycerol, 1.0% (w/v) DDM, 0.2% (w/v) CHS, 8 mM ATP, 40 µM CP-99,994/20 µM aprepitant/20 µM netupitant) followed by 30 CV of Wash Buffer II (50 mM HEPES pH 7.5, 500 mM NaCl, 15 mM imidazole, 10% (v/v) glycerol, 0.05% (w/v) DDM, 0.01% (w/v) CHS, 40 µM CP-99,994/20 µM aprepitant/20 µM netupitant). Antagonist-bound NK1R was eluted step-wise with four column volumes of Elution Buffer (50 mM HEPES pH 7.5, 500 mM NaCl, 250 mM imidazole, 10% (v/v) glycerol, 0.05% (w/v) DDM, 0.01% (w/v) CHS, 100 µM CP-99,994/50 µM aprepitant/50 µM netupitant). Protein-containing fractions were concentrated to 0.5 ml using a 100 kDa molecular weight cut-off Vivaspin 2 concentrator (Sartorius Stedim) and added to a PD MiniTrap G-25 column (GE Healthcare) equilibrated with G25 Buffer (50 mM HEPES pH 7.5, 500 mM NaCl, 10% (v/v) glycerol, 0.03% (w/v) DDM, 0.006% (w/v) CHS, 100 µM CP-99,994/50 µM aprepitant/50 µM netupitant) to remove imidazole. The complex was treated for 6 h with His-tagged 3C protease and PNGaseF (both prepared in-house) to remove the N-terminal affinity tags and deglycosylate the receptor. After incubation with Ni-NTA resin (GE Healthcare) overnight, cleaved receptor was collected as the flow-through and then concentrated to ~50–60 mg/ml with a 100 kDa molecular weight cut-off Vivaspin 2 concentrator. Protein concentrations were determined by absorbance at 280 nm on a Nanodrop 2000 spectrophotometer (Thermo Fisher Scientific). Protein purity and monodispersity were assessed by SDS-PAGE and analytical size-exclusion chromatography using a Sepax Nanofilm SEC-250 column.
Crystallisation in lipidic cubic phase
NK1R was crystallised using the in meso method at 20 °C. For this purpose, concentrated protein (~50–60 mg/ml) was mixed with molten monoolein (Sigma Aldrich) supplemented with 10% (w/w) cholesterol (Sigma Aldrich) using the twin-syringe method. The final protein:lipid ratio was 40:60 (v/v). 40 nl boli were dispensed on 96-well glass bases with a 120 µm spacer (SWISSCI), overlaid with 800 nl precipitant solution using a Gryphon LCP crystallisation robot (Art Robbins Instruments) and sealed with a cover glass. In initial screens of CP-99,994-bound NK1R, crystals appeared after less than 1 h in a broad range of conditions. Optimised crystals used for data collection were grown in a precipitant condition consisting of 100 mM MES pH 6.0, 31% (v/v) PEG400, 190–210 mM potassium acetate, 2.4% (v/v) 2,5-hexanediol and 50 µM CP-99,994. Aprepitant-bound NK1R yielded much fewer initial crystallisation hits compared to CP-99,994-bound NK1R. Optimised star-shaped crystals used for data collection of aprepitant-bound NK1R were obtained in a condition consisting of 100 mM sodium citrate pH 6.0, 31% (v/v) PEG400, 50–70 mM MgCl2 and 50 µM aprepitant. Crystals used for data collection of netupitant-bound NK1R were obtained in a condition consisting of 100 mM sodium citrate pH 6.0, 31% (v/v) PEG400, 40–50 mM Mg(HCO2)2 and 50 µM netupitant. Single crystals were mounted with Dual-Thickness MicroMounts (MiTeGen) of appropriate size for data collection and cryo-cooled in liquid nitrogen without the addition of further cryoprotectant.
Data collection and structure determination
X-ray diffraction data were collected at the X06SA beamline at the Swiss Light Source (SLS) of the Paul Scherrer Institute (PSI, Villigen, Switzerland) using a beam size of 10 × 10 μm and an EIGER 16 M detector. Datasets for CP-99,994-bound NK1R were collected using a beam attenuated to 10%, 0.1° of oscillation and 0.1 s exposure time. All other datasets were collected using a beam attenuated to 30%, 0.1° of oscillation and 0.05 s exposure time. Data from individual crystals were integrated using XDS66. Data merging and scaling was carried out using the program AIMLESS from the CCP4 suite67,68. Data collection statistics are reported in Table 1.
Initial phases were obtained by molecular replacement (MR) with the program Phaser69 using the truncated OX2R transmembrane domain (PDB ID 4S0V) and the separated PGS fusion protein31 as independent search models looking for one copy of each domain. Manual model building was performed in COOT70 using sigma-A weighted 2m|Fo|-|DFc|, m|Fo|-D|Fc| maps together with simulated-annealing and simple composite omit maps calculated using Phenix71. Initial refinement was carried out with REFMAC572 using maximum-likelihood restrained refinement in combination with the jelly-body protocol. Further and final stages of refinement were performed with Phenix.refine73 with positional, individual isotropic B-factor refinement and TLS. The final refinement statistics are presented in Table 1. Co-ordinates and structure factors have been deposited in the worldwide Protein Data Bank under accession codes 6HLL, 6HLO and 6HLP for the CP-99,994-, aprepitant- and netupitant-bound NK1R, respectively.
Whole-cell ligand-binding assay
HEK293T/17 cells (ATCC) were cultivated in Dulbecco’s modified medium (Sigma) supplemented with 100 units/ml penicillin, 100 µg/ml streptomycin (Sigma) and 10% (v/v) foetal calf serum (BioConcept). Cells were maintained at 37 °C in a humidified atmosphere of 5% CO2, 95% air. Transient transfections were performed with TransIT-293 (Mirus Bio) according to the manufacturer’s instructions.
Ligand-binding experiments were performed on whole HEK293T cells for comparison of affinities for wild-type and receptor mutants using a homogeneous time-resolved fluorescence (HTRF) binding assay. Receptor mutants were generated by site-directed mutagenesis and cloned into a mammalian expression vector (pcDNA3.1(+)) containing an N-terminal SNAP-tag (Cisbio). HEK293T cells were transiently transfected with receptor constructs and were seeded at 20,000 cells per well in poly-L-lysine-coated 384-well plates (Greiner). Forty-eight hours after transfection, cells were labelled with 50 nM SNAP-Lumi4-Tb (Cisbio) in assay buffer (20 mM HEPES pH 7.5, 100 mM NaCl, 3 mM MgCl2 and 0.2% (w/v) nonfat milk) for 1.5 h at 37 °C. Cells were washed four times with assay buffer and were then incubated for 2 h at RT in assay buffer containing fluorescently labelled peptide SP-HL488 (Substance P labelled with HiLyte Fluor 488 at Lys-3 (Anaspec))29. For competition binding, 20 nM of SP-HL488 tracer peptide and a concentration range of unlabelled antagonists as competitor were used. Fluorescence intensities were measured on an Infinite M1000 fluorescence plate reader (Tecan) with an excitation wavelength of 340 nm and emission wavelengths of 620 nm and 665 nm for Tb3+ and the fluorophore HiLyte Fluor 488, respectively. The ratio of FRET-donor and acceptor fluorescence intensities (F665 nm/F620 nm) was calculated. Total binding was obtained in the absence of competitor, and nonspecific binding was determined in the presence of 100 μM unlabelled competitor. Data were normalised to the specific binding for each individual experiment and were analysed by global fitting to a one-site heterologous competition equation with the GraphPad Prism software (version 6.07, GraphPad). To obtain Ki values, data were corrected for fluorescent ligand occupancy of each mutant with the Cheng-Prusoff equation as Ki = IC50/(1 + [fl. ligand]/Kd).
Reporting summary
Further information on experimental design is available in the Nature Research Reporting Summary linked to this article.
Data availability
Atomic coordinates and structure factors have been deposited in the Protein Data Bank under accession codes 6HLL, 6HLO and 6HLP for the CP-99,994-, aprepitant- and netupitant-bound NK1R, respectively. Data supporting the findings of this manuscript are available from the corresponding author upon reasonable request. The source data underlying Fig. 2 and Supplementary Table 1 are provided as a Source Data file.
References
Hökfelt, T., Pernow, B. & Wahren, J. Substance P: a pioneer amongst neuropeptides. J. Intern. Med. 249, 27–40 (2001).
Severini, C., Improta, G., Falconieri-Erspamer, G., Salvadori, S. & Erspamer, V. The tachykinin peptide family. Pharmacol. Rev. 54, 285–322 (2002).
Steinhoff, M. S. et al. Tachykinins and their receptors: contributions to physiological control and the mechanisms of disease. Physiol. Rev. 94, 265–301 (2014).
Helke, C. J., Krause, J. E., Mantyh, P. W., Couture, R. & Bannon, M. J. Diversity in mammalian tachykinin peptidergic neurons: multiple peptides, receptors, and regulatory mechanisms. FASEB J. 4, 1606–1615 (1990).
Mantyh, P. W., Gates, T., Mantyh, C. R. & Maggio, J. E. Autoradiographic localization and characterization of tachykinin receptor binding sites in the rat brain and peripheral tissues. J. Neurosci. 9, 258–279 (1989).
McLean, S. et al. Activity and distribution of binding sites in brain of a nonpeptide substance P (NK1) receptor antagonist. Science 251, 437–439 (1991).
Buck, S. H., Maurin, Y., Burks, T. F. & Yamamura, H. I. High-affinity 3H-substance P binding to longitudinal muscle membranes of the guinea pig small intestine. Life Sci. 34, 497–507 (1984).
Saito, R. et al. Pharmacological properties of the tachykinin receptor subtype in the endothelial cell and vasodilation. Ann. N. Y. Acad. Sci. 632, 457–459 (1991).
Payan, D. G., Brewster, D. R. & Goetzl, E. J. Stereospecific receptors for substance P on cultured human IM-9 lymphoblasts. J. Immunol. 133, 3260–3265 (1984).
Tattersall, F. D. et al. Tachykinin NK1 receptor antagonists act centrally to inhibit emesis induced by the chemotherapeutic agent cisplatin in ferrets. Neuropharmacology 35, 1121–1129 (1996).
Basbaum, A. I. et al. Primary afferent tachykinins are required to experience moderate to intense pain. Nature 392, 390–394 (1998).
Hunt, S. P. et al. Altered nociception, analgesia and aggression in mice lacking the receptor for substance P. Nature 392, 394–397 (1998).
Pintér, E., Pozsgai, G., Hajna, Z., Helyes, Z. & Szolcsányi, J. Neuropeptide receptors as potential drug targets in the treatment of inflammatory conditions. Br. J. Clin. Pharmacol. 77, 5–20 (2014).
Ständer, S., Siepmann, D., Herrgott, I., Sunderkötter, C. & Luger, T. A. Targeting the neurokinin receptor 1 with aprepitant: a novel antipruritic strategy. PLoS ONE 5, e10968 (2010).
Kramer, M. S. Distinct mechanism for antidepressant activity by blockade of central substance P receptors. Science 281, 1640–1645 (1998).
Ratti, E. et al. Full central neurokinin-1 receptor blockade is required for efficacy in depression: evidence from orvepitant clinical studies. J. Psychopharmacol. 27, 424–434 (2013).
Hauser, A. S., Attwood, M. M., Rask-Andersen, M., Schiöth, H. B. & Gloriam, D. E. Trends in GPCR drug discovery: new agents, targets and indications. Nat. Rev. Drug. Discov. 16, 829–842 (2017).
Jensen, D. D. et al. Neurokinin 1 receptor signaling in endosomes mediates sustained nociception and is a viable therapeutic target for prolonged pain relief. Sci. Transl. Med. 9, eaal3447 (2017).
Snider, R. et al. A potent nonpeptide antagonist of the substance P (NK1) receptor. Science 251, 435–437 (1991).
Desai, M. C., Lefkowitz, S. L., Thadeio, P. F., Longo, K. P. & Snider, R. M. Discovery of a potent substance P antagonist: recognition of the key molecular determinant. J. Med. Chem. 35, 4911–4913 (1992).
McLean, S. et al. Pharmacology of CP-99,994; a nonpeptide antagonist of the tachykinin neurokinin-1 receptor. J. Pharmacol. Exp. Ther. 267, 472–479 (1993).
Hale, J. J. et al. 2(S)-((3,5-Bis(trifluoromethyl)benzyl)oxy)-3(S)-phenyl-4-((3-oxo-1,2,4-triazol- 5-yl)methyl)morpholine (1): a potent, orally active, morpholine-based human neurokinin-1 receptor antagonist. J. Med. Chem. 39, 1760–1762 (1996).
Cascieri, M. A. et al. Characterization of the binding and activity of a high affinity, pseudoirreversible morpholino tachykinin NK1 receptor antagonist. Eur. J. Pharmacol. 325, 253–261 (1997).
Hale, J. J. et al. Structural optimization affording 2-(R)-(1-(R)-3, 5-bis(trifluoromethyl)phenylethoxy)-3-(S)-(4-fluoro)phenyl-4- (3-oxo-1,2,4-triazol-5-yl)methylmorpholine, a potent, orally active, long-acting morpholine acetal human NK-1 receptor antagonist. J. Med. Chem. 41, 4607–4614 (1998).
Hargreaves, R. et al. Development of aprepitant, the first neurokinin-1 receptor antagonist for the prevention of chemotherapy-induced nausea and vomiting. Ann. N. Y. Acad. Sci. 1222, 40–48 (2011).
Fong, T. M., Huang, R. R. & Strader, C. D. Localization of agonist and antagonist binding domains of the human neurokinin-1 receptor. J. Biol. Chem. 267, 25664–25667 (1992).
Gether, U. et al. Different binding epitopes on the NK1 receptor for substance P and a non-peptide antagonist. Nature 362, 345–348 (1993).
Turcatti, G. et al. Characterization of non-peptide antagonist and peptide agonist binding sites of the NK1 receptor with fluorescent ligands. J. Biol. Chem. 272, 21167–21175 (1997).
Schütz, M. et al. Directed evolution of G protein-coupled receptors in yeast for higher functional production in eukaryotic expression hosts. Sci. Rep. 6, 21508 (2016).
Ballesteros, J. A. & Weinstein, H. Integrated methods for the construction of three-dimensional models and computational probing of structure-function relations in G protein-coupled receptors, Methods in Neurosciences 25, 366–428 (Academic Press, 1995).
Yin, J., Mobarec, J. C., Kolb, P. & Rosenbaum, D. M. Crystal structure of the human OX2 orexin receptor bound to the insomnia drug suvorexant. Nature 519, 247–250 (2015).
Yang, Z. et al. Structural basis of ligand binding modes at the neuropeptide Y Y1 receptor. Nature 556, 520–524 (2018).
Shihoya, W. et al. X-ray structures of endothelin ETB receptor bound to clinical antagonist bosentan and its analog. Nat. Struct. Mol. Biol. 24, 758–764 (2017).
Preininger, A. M., Meiler, J. & Hamm, H. E. Conformational flexibility and structural dynamics in GPCR-mediated G protein activation: a perspective. J. Mol. Biol. 425, 2288–2298 (2013).
Greenfeder, S. et al. Two related neurokinin-1 receptor antagonists have overlapping but different binding sites. Bioorg. Med. Chem. 6, 189–194 (1998).
Holst, B., Zoffmann, S., Elling, C. E., Hjorth, S. A. & Schwartz, T. W. Steric hindrance mutagenesis versus alanine scan in mapping of ligand binding sites in the tachykinin NK1 receptor. Mol. Pharmacol. 53, 166–175 (1998).
Thompson, C. A. Netupitant–palonosetron combination approved by FDA. Am. J. Health Syst. Pharm. 71, 2000 (2014).
Hoffmann, T. et al. Design and synthesis of a novel, achiral class of highly potent and selective, orally active neurokinin-1 receptor antagonists. Bioorg. Med. Chem. Lett. 16, 1362–1365 (2006).
Elling, C. E., Nielsen, S. M. & Schwartz, T. W. Conversion of antagonist-binding site to metal-ion site in the tachykinin NK-1 receptor. Nature 374, 74–77 (1995).
Warne, T. et al. The structural basis for agonist and partial agonist action on a β1-adrenergic receptor. Nature 469, 241–244 (2011).
Ring, A. M. et al. Adrenaline-activated structure of β2-adrenoceptor stabilized by an engineered nanobody. Nature 502, 575–579 (2013).
Bondar, A.-N. & White, S. H. Hydrogen bond dynamics in membrane protein function. Biochim. Biophys. Acta 1818, 942–950 (2012).
Engelman, D. M., Zhou, F. X., Cocco, M. J., Russ, W. P. & Brunger, A. T. Interhelical hydrogen bonding drives strong interactions in membrane proteins. Nat. Struct. Biol. 7, 154–160 (2000).
Patel, A. B. et al. Changes in interhelical hydrogen bonding upon rhodopsin activation. J. Mol. Biol. 347, 803–812 (2005).
Rasmussen, S. G. F. et al. Crystal structure of the β2 adrenergic receptor-Gs protein complex. Nature 477, 549–555 (2011).
Che, T. et al. Structure of the nanobody-stabilized active state of the kappa opioid receptor. Cell 172, 55–67 (2018). e15.
Fierens, F. L. P., Vanderheyden, P. M. L., De Backer, J.-P. & Vauquelin, G. Insurmountable angiotensin AT1 receptor antagonists: the role of tight antagonist binding. Eur. J. Pharmacol. 372, 199–206 (1999).
Vauquelin, G., Morsing, P., Fierens, F. L. P., De Backer, J.-P. & Vanderheyden, P. M. L. A two-state receptor model for the interaction between angiotensin II type 1 receptors and non-peptide antagonists. Biochem. Pharmacol. 61, 277–284 (2001).
Lindström, E. et al. Neurokinin 1 receptor antagonists: correlation between in vitro receptor interaction and in vivo efficacy. J. Pharmacol. Exp. Ther. 322, 1286–1293 (2007).
Rizzi, A. et al. In vitro and in vivo pharmacological characterization of the novel NK1 receptor selective antagonist Netupitant. Peptides 37, 86–97 (2012).
Katritch, V., Cherezov, V. & Stevens, R. C. Structure-function of the G protein–coupled receptor superfamily. Annu. Rev. Pharmacol. Toxicol. 53, 531–556 (2013).
Nygaard, R., Valentin-Hansen, L., Mokrosinski, J., Frimurer, T. M. & Schwartz, T. W. Conserved water-mediated hydrogen bond network between TM-I, -II, -VI, and -VII in 7TM receptor activation. J. Biol. Chem. 285, 19625–19636 (2010).
Gutiérrez-de-Terán, H. et al. The role of a sodium ion binding site in the allosteric modulation of the A2A adenosine G protein-coupled receptor. Structure 21, 2175–2185 (2013).
Katritch, V. et al. Allosteric sodium in class A GPCR signaling. Trends Biochem. Sci. 39, 233–244 (2014).
Mirzadegan, T., Benkö, G., Filipek, S. & Palczewski, K. Sequence analyses of G-protein-coupled receptors: similarities to rhodopsin. Biochemistry 42, 2759–2767 (2003).
Valentin-Hansen, L., Frimurer, T. M., Mokrosinski, J., Holliday, N. D. & Schwartz, T. W. Biased Gs versus Gq proteins and β-arrestin signaling in the NK1 receptor determined by interactions in the water hydrogen bond network. J. Biol. Chem. 290, 24495–24508 (2015).
Holst, B., Hastrup, H., Raffetseder, U., Martini, L. & Schwartz, T. W. Two active molecular phenotypes of the tachykinin NK1 receptor revealed by G-protein fusions and mutagenesis. J. Biol. Chem. 276, 19793–19799 (2001).
Wang, S. et al. D4 dopamine receptor high-resolution structures enable the discovery of selective agonists. Science 358, 381–386 (2017).
Fenalti, G. et al. Molecular control of δ-opioid receptor signalling. Nature 506, 191–196 (2014).
Miller-Gallacher, J. L. et al. The 2.1 Å resolution structure of cyanopindolol-bound β1-adrenoceptor identifies an intramembrane Na+ ion that stabilises the ligand-free receptor. PLoS ONE 9, e92727 (2014).
Liu, W. et al. Structural basis for allosteric regulation of GPCRs by sodium ions. Science 337, 232–236 (2012).
Dawson, L. A. & Smith, P. W. Therapeutic utility of NK3 receptor antagonists for the treatment of schizophrenia. Curr. Pharm. Des. 16, 344–357 (2010).
Spooren, W., Riemer, C. & Meltzer, H. NK3 receptor antagonists: the next generation of antipsychotics? Nat. Rev. Drug. Discov. 4, 967–975 (2005).
Prague, J. K. et al. Neurokinin 3 receptor antagonism as a novel treatment for menopausal hot flushes: a phase 2, randomised, double-blind, placebo-controlled trial. Lancet 389, 1809–1820 (2017).
Alexandrov, A. I., Mileni, M., Chien, E. Y. T., Hanson, M. A. & Stevens, R. C. Microscale fluorescent thermal stability assay for membrane proteins. Structure 16, 351–359 (2008).
Kabsch, W. Integration, scaling, space‐group assignment and post‐refinement. Acta Crystallogr. D Biol. Crystallogr. 66, 133–144 (2010).
Dodson, E. J., Winn, M. & Ralph, A. Collaborative Computational Project, Number 4. ‘The CCP4 Suite: Programs for Protein Crystallography’. Acta Crystallogr. D Biol. Crystallogr. 50, 760–763 (1994).
Evans, P. R. & Murshudov, G. N. How good are my data and what is the resolution? Acta Crystallogr. D Biol. Crystallogr. 69, 1204–1214 (2013).
McCoy, A. J. et al. Phaser crystallographic software. J. Appl. Crystallogr. 40, 658–674 (2007).
Emsley, P., Lohkamp, B., Scott, W. G. & Cowtan, K. IUCr. Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr. 66, 486–501 (2010).
Adams, P. D. et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 66, 213–221 (2010).
Murshudov, G. N. et al. REFMAC5 for the refinement of macromolecular crystal structures. Acta Crystallogr. D Biol. Crystallogr. 67, 355–367 (2011).
Afonine, P. V. et al. Towards automated crystallographic structure refinement with phenix.refine. Acta Crystallogr. D Biol. Crystallogr. 68, 352–367 (2012).
Acknowledgements
We thank B. Blattmann of the Protein Crystallisation Center at the University of Zurich, the staff of the X06SA beamline at the Paul Scherrer Institute for support during data collection and I. Berger at the European Molecular Biology Laboratory for providing us with baculovirus transfer vectors. We thank G. Meier and B. Aebli for support during protein production. We thank M. Hillenbrand for support during initial pharmacological characterisation. We would furthermore like to thank F. Zosel for critical reading of the manuscript. C.K. is the recipient of a fellowship of the German Academy of Sciences Leopoldina (LPDS 2009-48) and a Marie Curie fellowship of the European Commission (FP7-PEOPLE-2011-IEF #299208). This work was supported by Schweizerischer Nationalfonds Grants 31003A_153143, 31003A_182334 and KTI grant 18022.1 PFLS-LS, all to A.P.
Author information
Authors and Affiliations
Contributions
J.S. devised and carried out the mutagenesis and thermostabilisation of the receptor, designed and characterised crystallisation constructs, expressed, purified, crystallised the NK1R-PGS fusion protein and harvested crystals. J.S. established the receptor purification and LCP crystallisation platforms. J.E. supported cloning, expression and crystal harvesting. J.S., J.E. and A.S.D. collected data. J.S., J.E., P.R. and A.S.D. processed the data, solved and refined the structures. C.K. performed ligand-binding experiments and analysed the data. M.S. performed directed evolution in yeast. Project management was carried out by J.S. and A.P. The manuscript was prepared by J.S., J.E., A.S.D. and A.P. All authors contributed to the final editing and approval of the manuscript.
Corresponding author
Ethics declarations
Competing interests
P.R., M.S. and A.S.D. are employees of Sosei Heptares, a company with activities in the GPCR field. All other authors declare no competing interests.
Additional information
Journal peer review information: Nature Communications thanks the anonymous reviewers for their contribution to the peer review of this work. [Peer reviewer reports are available.]
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Schöppe, J., Ehrenmann, J., Klenk, C. et al. Crystal structures of the human neurokinin 1 receptor in complex with clinically used antagonists. Nat Commun 10, 17 (2019). https://doi.org/10.1038/s41467-018-07939-8
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41467-018-07939-8
This article is cited by
-
Fluorine-19 labeling of the tryptophan residues in the G protein-coupled receptor NK1R using the 5-fluoroindole precursor in Pichia pastoris expression
Journal of Biomolecular NMR (2024)
-
A Vaccinia-based system for directed evolution of GPCRs in mammalian cells
Nature Communications (2023)
-
Structure-based design of bitopic ligands for the µ-opioid receptor
Nature (2023)
-
Selective G protein signaling driven by substance P–neurokinin receptor dynamics
Nature Chemical Biology (2022)
-
Neuropeptidergic regulation of compulsive ethanol seeking in C. elegans
Scientific Reports (2022)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.