Abstract
The deubiquitylating enzymes (DUBs, also known as deubiquitylases or deubiquitinases) maintain the dynamic state of the cellular ubiquitome by releasing conjugated ubiquitin from proteins. In light of the many cellular functions of ubiquitin, DUBs occupy key roles in almost all aspects of cell behaviour. Many DUBs show selectivity for particular ubiquitin linkage types or positions within ubiquitin chains. Others show chain-type promiscuity but can select a distinct palette of protein substrates via specific protein–protein interactions established through binding modules outside of the catalytic domain. The ubiquitin chain cleavage mode or chain linkage specificity has been related directly to biological functions. Examples include regulation of protein degradation and ubiquitin recycling by the proteasome, DNA repair pathways and innate immune signalling. DUB cleavage specificity is also being harnessed for analysis of ubiquitin chain architecture that is assembled on specific proteins. The recent development of highly specific DUB inhibitors heralds their emergence as a new class of therapeutic targets for numerous diseases.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$189.00 per year
only $15.75 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
Change history
19 February 2019
Figure 2 of the article as originally published contained a graphic editing error, whereby the publisher’s redrawn figure wrongly indicated the presence of a Drosophila melanogaster orthologue of ZUP1. This has been corrected in the HTML and PDF versions of the manuscript.
References
Komander, D. & Rape, M. The ubiquitin code. Annu. Rev. Biochem. 81, 203–229 (2012).
Clague, M. J., Heride, C. & Urbe, S. The demographics of the ubiquitin system. Trends Cell Biol. 25, 417–426 (2015).
Swatek, K. N. & Komander, D. Ubiquitin modifications. Cell Res. 26, 399–422 (2016).
Yau, R. & Rape, M. The increasing complexity of the ubiquitin code. Nat. Cell Biol. 18, 579–586 (2016).
Grou, C. P., Pinto, M. P., Mendes, A. V., Domingues, P. & Azevedo, J. E. The de novo synthesis of ubiquitin: identification of deubiquitinases acting on ubiquitin precursors. Sci. Rep. 5, 12836 (2015).
Walczak, H., Iwai, K. & Dikic, I. Generation and physiological roles of linear ubiquitin chains. BMC Biol. 10, 23 (2012).
Rahighi, S. & Dikic, I. Selectivity of the ubiquitin-binding modules. FEBS Lett. 586, 2705–2710 (2012).
Kaiser, S. E. et al. Protein standard absolute quantification (PSAQ) method for the measurement of cellular ubiquitin pools. Nat. Methods 8, 691–696 (2011).
Komander, D., Clague, M. J. & Urbe, S. Breaking the chains: structure and function of the deubiquitinases. Nat. Rev. Mol. Cell Biol. 10, 550–563 (2009).
Clague, M. J. et al. Deubiquitylases from genes to organism. Physiol. Rev. 93, 1289–1315 (2013).
Mevissen, T. E. T. & Komander, D. Mechanisms of deubiquitinase specificity and regulation. Annu. Rev. Biochem. 86, 159–192 (2017).
Walden, M., Masandi, S. K., Pawlowski, K. & Zeqiraj, E. Pseudo-DUBs as allosteric activators and molecular scaffolds of protein complexes. Biochem. Soc. Trans. 46, 453–466 (2018).
Abdul Rehman, S. A. et al. MINDY-1 is a member of an evolutionarily conserved and structurally distinct new family of deubiquitinating enzymes. Mol. Cell 63, 146–155 (2016). This study identifies a new family of DUBs (MINDY) selective for Lys48-linked ubiquitin chains and presents a structural characterization of the catalytic domain of MINDY1.
Kristariyanto, Y. A., Abdul Rehman, S. A., Weidlich, S., Knebel, A. & Kulathu, Y. A single MIU motif of MINDY-1 recognizes K48-linked polyubiquitin chains. EMBO Rep. 18, 392–402 (2017).
Haahr, P. et al. ZUFSP deubiquitylates K63-linked polyubiquitin chains to promote genome stability. Mol. Cell 70, 165–174 (2018).
Hermanns, T. et al. A family of unconventional deubiquitinases with modular chain specificity determinants. Nat. Commun. 9, 799 (2018).
Hewings, D. S. et al. Reactive-site-centric chemoproteomics identifies a distinct class of deubiquitinase enzymes. Nat. Commun. 9, 1162 (2018).
Kwasna, D. et al. Discovery and characterization of ZUFSP/ZUP1, a distinct deubiquitinase class important for genome stability. Mol. Cell 70, 150–164 (2018). References 15–18 report the discovery of the ZUP1 family of DUBs and link the single human representative to Lys63 ubiquitin chain processing and maintenance of genome stability.
Winborn, B. J. et al. The deubiquitinating enzyme ataxin-3, a polyglutamine disease protein, edits Lys63 linkages in mixed linkage ubiquitin chains. J. Biol. Chem. 283, 26436–26443 (2008).
Mevissen, T. E. et al. OTU deubiquitinases reveal mechanisms of linkage specificity and enable ubiquitin chain restriction analysis. Cell 154, 169–184 (2013). This paper provides a systematic biochemical and structural survey of ubiquitin chain linkage specificity within the OTU family.
Schulz, S. et al. Ubiquitin-specific protease-like 1 (USPL1) is a SUMO isopeptidase with essential, non-catalytic functions. EMBO Rep. 13, 930–938 (2012).
Malakhov, M. P., Malakhova, O. A., Kim, K. I., Ritchie, K. J. & Zhang, D. E. UBP43 (USP18) specifically removes ISG15 from conjugated proteins. J. Biol. Chem. 277, 9976–9981 (2002).
Cavadini, S. et al. Cullin-RING ubiquitin E3 ligase regulation by the COP9 signalosome. Nature 531, 598–603 (2016).
Mevissen, T. E. T. et al. Molecular basis of Lys11-polyubiquitin specificity in the deubiquitinase Cezanne. Nature 538, 402–405 (2016).
Faesen, A. C. et al. The differential modulation of USP activity by internal regulatory domains, interactors and eight ubiquitin chain types. Chem. Biol. 18, 1550–1561 (2011). This paper presents a biochemical comparison of 12 USP family members that provides enzyme kinetic constants and characterizes their respective selectivity towards eight ubiquitin chain linkage types.
Ritorto, M. S. et al. Screening of DUB activity and specificity by MALDI-TOF mass spectrometry. Nat. Commun. 5, 4763 (2014). This paper presents a novel matrix-assisted laser desorption/ionization time of flight (MALDI-TOF) assay for screening DUB activity and a survey of published DUB inhibitors that generally indicate poor specificity.
Sato, Y. et al. Structures of CYLD USP with Met1- or Lys63-linked diubiquitin reveal mechanisms for dual specificity. Nat. Struct. Mol. Biol. 22, 222–229 (2015).
Gersch, M. et al. Mechanism and regulation of the Lys6-selective deubiquitinase USP30. Nat. Struct. Mol. Biol. 24, 920–930 (2017).
Sato, Y. et al. Structural basis for specific cleavage of Lys6-linked polyubiquitin chains by USP30. Nat. Struct. Mol. Biol. 24, 911–919 (2017).
Damgaard, R. B. et al. The deubiquitinase OTULIN is an essential negative regulator of inflammation and autoimmunity. Cell 166, 1215–1230 (2016).
Urbe, S. et al. Systematic survey of deubiquitinase localisation identifies USP21 as a regulator of centrosome and microtubule associated functions. Mol. Biol. Cell 23, 1095–1103 (2012). This paper details the subcellular localization map for >60 GFP-tagged DUBs.
Hassink, G. C. et al. The ER-resident ubiquitin-specific protease 19 participates in the UPR and rescues ERAD substrates. EMBO Rep. 10, 755–761 (2009).
Marcassa, E. et al. Dual role of USP30 in controlling basal pexophagy and mitophagy. EMBO Rep. 19, e45595 (2018).
Nishi, R. et al. Systematic characterization of deubiquitylating enzymes for roles in maintaining genome integrity. Nat. Cell Biol. 16, 1016–1026 (2014).
Itzhak, D. N., Tyanova, S., Cox, J. & Borner, G. H. Global, quantitative and dynamic mapping of protein subcellular localization. eLife 5, e16950 (2016).
Li, J. et al. USP33 regulates centrosome biogenesis via deubiquitination of the centriolar protein CP110. Nature 495, 255–259 (2013).
Heride, C. et al. The centrosomal deubiquitylase USP21 regulates Gli1 transcriptional activity and stability. J. Cell Sci. 129, 4001–4013 (2016).
Li, X. et al. USP9X regulates centrosome duplication and promotes breast carcinogenesis. Nat. Commun. 8, 14866 (2017).
Wang, Q. et al. The X-linked deubiquitinase USP9X is an integral component of centrosome. J. Biol. Chem. 292, 12874–12884 (2017).
Lu, Y. et al. USP19 deubiquitinating enzyme supports cell proliferation by stabilizing KPC1, a ubiquitin ligase for p27Kip1. Mol. Cell. Biol. 29, 547–558 (2009).
Thorne, C., Eccles, R. L., Coulson, J. M., Urbe, S. & Clague, M. J. Isoform-specific localization of the deubiquitinase USP33 to the Golgi apparatus. Traffic 12, 1563–1574 (2011).
Leznicki, P. et al. Expansion of DUB functionality generated by alternative isoforms — USP35, a case study. J. Cell Sci. 131, jcs212753 (2018).
Wang, Y. et al. Deubiquitinating enzymes regulate PARK2-mediated mitophagy. Autophagy 11, 595–606 (2015).
Hart, T. et al. Evaluation and design of genome-wide CRISPR/SpCas9 knockout screens. G3 7, 2719–2727 (2017).
Meyers, R. M. et al. Computational correction of copy number effect improves specificity of CRISPR-Cas9 essentiality screens in cancer cells. Nat. Genet. 49, 1779–1784 (2017).
Pathare, G. R. et al. Crystal structure of the proteasomal deubiquitylation module Rpn8-Rpn11. Proc. Natl Acad. Sci. USA 111, 2984–2989 (2014).
Reyes-Turcu, F. E. et al. The ubiquitin binding domain ZnF UBP recognizes the C-terminal diglycine motif of unanchored ubiquitin. Cell 124, 1197–1208 (2006).
Bonnet, J., Romier, C., Tora, L. & Devys, D. Zinc-finger UBPs: regulators of deubiquitylation. Trends Biochem. Sci. 33, 369–375 (2008).
Xie, X. et al. Deubiquitylases USP5 and USP13 are recruited to and regulate heat-induced stress granules through their deubiquitylating activities. J. Cell Sci. 131, jcs.210856 (2018).
Richardson, L. A. et al. A conserved deubiquitinating enzyme controls cell growth by regulating RNA polymerase I stability. Cell Rep. 2, 372–385 (2012).
Hutten, S., Chachami, G., Winter, U., Melchior, F. & Lamond, A. I. A role for the Cajal-body-associated SUMO isopeptidase USPL1 in snRNA transcription mediated by RNA polymerase II. J. Cell Sci. 127, 1065–1078 (2014).
Ciechanover, A. Proteolysis: from the lysosome to ubiquitin and the proteasome. Nat. Rev. Mol. Cell. Biol. 6, 79–86 (2005).
Meyer, H. J. & Rape, M. Enhanced protein degradation by branched ubiquitin chains. Cell 157, 910–921 (2014).
Lu, Y., Lee, B. H., King, R. W., Finley, D. & Kirschner, M. W. Substrate degradation by the proteasome: a single-molecule kinetic analysis. Science 348, 1250834 (2015).
Yu, H. & Matouschek, A. Recognition of client proteins by the proteasome. Annu. Rev. Biophys. 46, 149–173 (2017).
Ohtake, F., Tsuchiya, H., Saeki, Y. & Tanaka, K. K63 ubiquitylation triggers proteasomal degradation by seeding branched ubiquitin chains. Proc. Natl Acad. Sci. USA 115, E1401–E1408 (2018).
de Poot, S. A. H., Tian, G. & Finley, D. Meddling with fate: the proteasomal deubiquitinating enzymes. J. Mol. Biol. 429, 3525–3545 (2017).
Beck, F. et al. Near-atomic resolution structural model of the yeast 26S proteasome. Proc. Natl Acad. Sci. USA 109, 14870–14875 (2012).
Matyskiela, M. E., Lander, G. C. & Martin, A. Conformational switching of the 26S proteasome enables substrate degradation. Nat. Struct. Mol. Biol. 20, 781–788 (2013).
Worden, E. J., Padovani, C. & Martin, A. Structure of the Rpn11-Rpn8 dimer reveals mechanisms of substrate deubiquitination during proteasomal degradation. Nat. Struct. Mol. Biol. 21, 220–227 (2014).
Dambacher, C. M., Worden, E. J., Herzik, M. A., Martin, A. & Lander, G. C. Atomic structure of the 26S proteasome lid reveals the mechanism of deubiquitinase inhibition. eLife 5, e13027 (2016).
Worden, E. J., Dong, K. C. & Martin, A. An AAA motor-driven mechanical switch in Rpn11 controls deubiquitination at the 26S proteasome. Mol. Cell 67, 799–811 (2017). This structural study indicates a rate-limiting conformational switch in the proteasomal DUB Rpn11 upon ubiquitin binding that is accelerated by mechanical translocation of a proteasomal substrate driven by AAA+ motor proteins.
Verma, R. et al. Role of Rpn11 metalloprotease in deubiquitination and degradation by the 26S proteasome. Science 298, 611–615 (2002).
Yao, T. & Cohen, R. E. A cryptic protease couples deubiquitination and degradation by the proteasome. Nature 419, 403–407 (2002).
Sato, Y. et al. Structural basis for specific cleavage of Lys 63-linked polyubiquitin chains. Nature 455, 358–362 (2008).
Koulich, E., Li, X. & Demartino, G. N. Relative structural and functional roles of multiple deubiquitylating proteins associated with mammalian 26S proteasome. Mol. Biol. Cell 19, 1072–1082 (2008).
Sahtoe, D. D. et al. Mechanism of UCH-L5 activation and inhibition by DEUBAD domains in RPN13 and INO80G. Mol. Cell 57, 887–900 (2015).
Lee, B. H. et al. USP14 deubiquitinates proteasome-bound substrates that are ubiquitinated at multiple sites. Nature 532, 398–401 (2016). This biochemical study reveals en bloc removal of supernumerary ubiquitin chains by the proteasomal DUB USP14.
Shi, Y. et al. Rpn1 provides adjacent receptor sites for substrate binding and deubiquitination by the proteasome. Science 351, aad9421 (2016).
VanderLinden, R. T. et al. Structural basis for the activation and inhibition of the UCH37 deubiquitylase. Mol. Cell 61, 487 (2016).
Kirkpatrick, D. S. et al. Quantitative analysis of in vitro ubiquitinated cyclin B1 reveals complex chain topology. Nat. Cell Biol. 8, 700–710 (2006).
Zhou, Z. R., Zhang, Y. H., Liu, S., Song, A. X. & Hu, H. Y. Length of the active-site crossover loop defines the substrate specificity of ubiquitin C-terminal hydrolases for ubiquitin chains. Biochem. J. 441, 143–149 (2012).
Stone, M. et al. Uch2/Uch37 is the major deubiquitinating enzyme associated with the 26S proteasome in fission yeast. J. Mol. Biol. 344, 697–706 (2004).
Hamazaki, J. et al. A novel proteasome interacting protein recruits the deubiquitinating enzyme UCH37 to 26S proteasomes. EMBO J. 25, 4524–4536 (2006).
Lam, Y. A., Xu, W., DeMartino, G. N. & Cohen, R. E. Editing of ubiquitin conjugates by an isopeptidase in the 26S proteasome. Nature 385, 737–740 (1997).
Yao, T. et al. Distinct modes of regulation of the Uch37 deubiquitinating enzyme in the proteasome and in the Ino80 chromatin-remodeling complex. Mol. Cell 31, 909–917 (2008).
Lenoir, W. F., Lim, T. L. & Hart, T. PICKLES: the database of pooled in-vitro CRISPR knockout library essentiality screens. Nucleic Acids Res. 46, D776–D780 (2018).
Sanchez-Pulido, L., Kong, L. & Ponting, C. P. A common ancestry for BAP1 and Uch37 regulators. Bioinformatics 28, 1953–1956 (2012).
Sahtoe, D. D., van Dijk, W. J., Ekkebus, R., Ovaa, H. & Sixma, T. K. BAP1/ASXL1 recruitment and activation for H2A deubiquitination. Nat. Commun. 7, 10292 (2016).
Seale, R. L. Rapid turnover of the histone-ubiquitin conjugate, protein A24. Nucleic Acids Res. 9, 3151–3158 (1981).
Joo, H. Y. et al. Regulation of cell cycle progression and gene expression by H2A deubiquitination. Nature 449, 1068–1072 (2007).
Zhu, P. et al. A histone H2A deubiquitinase complex coordinating histone acetylation and H1 dissociation in transcriptional regulation. Mol. Cell 27, 609–621 (2007).
Zhang, X. Y., Pfeiffer, H. K., Thorne, A. W. & McMahon, S. B. USP22, an hSAGA subunit and potential cancer stem cell marker, reverses the polycomb-catalyzed ubiquitylation of histone H2A. Cell Cycle 7, 1522–1524 (2008).
Scheuermann, J. C. et al. Histone H2A deubiquitinase activity of the Polycomb repressive complex PR-DUB. Nature 465, 243–247 (2010).
Morgan, M. T. et al. Structural basis for histone H2B deubiquitination by the SAGA DUB module. Science 351, 725–728 (2016).
Abdel-Rahman, M. H. et al. Germline BAP1 mutation predisposes to uveal melanoma, lung adenocarcinoma, meningioma, and other cancers. J. Med. Genet. 48, 856–859 (2011).
Daou, S. et al. The BAP1/ASXL2 histone H2A deubiquitinase complex regulates cell proliferation and is disrupted in cancer. J. Biol. Chem. 290, 28643–28663 (2015).
Kee, Y. & Huang, T. T. Role of deubiquitinating enzymes in DNA repair. Mol. Cell. Biol. 36, 524–544 (2016).
Nijman, S. M. et al. The deubiquitinating enzyme USP1 regulates the Fanconi anemia pathway. Mol. Cell 17, 331–339 (2005).
Huang, T. T. et al. Regulation of monoubiquitinated PCNA by DUB autocleavage. Nat. Cell Biol. 8, 339–347 (2006).
Elia, A. E. et al. Quantitative proteomic atlas of ubiquitination and acetylation in the DNA damage response. Mol. Cell 59, 867–881 (2015).
Moyal, L. et al. Requirement of ATM-dependent monoubiquitylation of histone H2B for timely repair of DNA double-strand breaks. Mol. Cell 41, 529–542 (2011).
Nakamura, K. et al. Regulation of homologous recombination by RNF20-dependent H2B ubiquitination. Mol. Cell 41, 515–528 (2011).
Fuchs, G. & Oren, M. Writing and reading H2B monoubiquitylation. Biochim. Biophys. Acta 1839, 694–701 (2014).
Ramachandran, S. et al. The SAGA deubiquitination module promotes DNA repair and class switch recombination through ATM and DNAPK-mediated gammaH2AX formation. Cell Rep. 15, 1554–1565 (2016).
Thorslund, T. et al. Histone H1 couples initiation and amplification of ubiquitin signalling after DNA damage. Nature 527, 389–393 (2015).
Lee, B. L., Singh, A., Mark Glover, J. N., Hendzel, M. J. & Spyracopoulos, L. Molecular basis for K63-linked ubiquitination processes in double-strand DNA break repair: a focus on kinetics and dynamics. J. Mol. Biol. 429, 3409–3429 (2017).
Nowsheen, S. et al. L3MBTL2 orchestrates ubiquitin signalling by dictating the sequential recruitment of RNF8 and RNF168 after DNA damage. Nat. Cell Biol. 20, 455–464 (2018).
Mattiroli, F. et al. RNF168 ubiquitinates K13-15 on H2A/H2AX to drive DNA damage signaling. Cell 150, 1182–1195 (2012).
Nakada, S. et al. Non-canonical inhibition of DNA damage-dependent ubiquitination by OTUB1. Nature 466, 941–946 (2010).
Juang, Y. C. et al. OTUB1 co-opts Lys48-linked ubiquitin recognition to suppress E2 enzyme function. Mol. Cell 45, 384–397 (2012).
Wiener, R., Zhang, X., Wang, T. & Wolberger, C. The mechanism of OTUB1-mediated inhibition of ubiquitination. Nature 483, 618–622 (2012).
Sy, S. M. et al. The ubiquitin specific protease USP34 promotes ubiquitin signaling at DNA double-strand breaks. Nucleic Acids Res. 41, 8572–8580 (2013).
Fradet-Turcotte, A. et al. 53BP1 is a reader of the DNA-damage-induced H2A Lys 15 ubiquitin mark. Nature 499, 50–54 (2013).
Wang, Z. et al. USP51 deubiquitylates H2AK13,15ub and regulates DNA damage response. Genes Dev. 30, 946–959 (2016).
Paul, A. & Wang, B. RNF8- and Ube2S-dependent ubiquitin lysine 11-linkage modification in response to DNA damage. Mol. Cell 66, 458–472 (2017).
Zheng, H. et al. A BRISC-SHMT complex deubiquitinates IFNAR1 and regulates interferon responses. Cell Rep. 5, 180–193 (2013).
Hashizume, R. et al. The RING heterodimer BRCA1-BARD1 is a ubiquitin ligase inactivated by a breast cancer-derived mutation. J. Biol. Chem. 276, 14537–14540 (2001).
Densham, R. M. et al. Human BRCA1-BARD1 ubiquitin ligase activity counteracts chromatin barriers to DNA resection. Nat. Struct. Mol. Biol. 23, 647–655 (2016).
Uckelmann, M. et al. USP48 restrains resection by site-specific cleavage of the BRCA1 ubiquitin mark from H2A. Nat. Commun. 9, 229 (2018).
Orthwein, A. et al. A mechanism for the suppression of homologous recombination in G1 cells. Nature 528, 422–426 (2015).
Deng, L. et al. Activation of the IkappaB kinase complex by TRAF6 requires a dimeric ubiquitin-conjugating enzyme complex and a unique polyubiquitin chain. Cell 103, 351–361 (2000).
Jiang, X. & Chen, Z. J. The role of ubiquitylation in immune defence and pathogen evasion. Nat. Rev. Immunol. 12, 35–48 (2011).
Haas, T. L. et al. Recruitment of the linear ubiquitin chain assembly complex stabilizes the TNF-R1 signaling complex and is required for TNF-mediated gene induction. Mol. Cell 36, 831–844 (2009).
Damgaard, R. B. et al. The ubiquitin ligase XIAP recruits LUBAC for NOD2 signaling in inflammation and innate immunity. Mol. Cell 46, 746–758 (2012).
Ikeda, F. et al. SHARPIN forms a linear ubiquitin ligase complex regulating NF-kappaB activity and apoptosis. Nature 471, 637–641 (2011).
Emmerich, C. H. et al. Activation of the canonical IKK complex by K63/M1-linked hybrid ubiquitin chains. Proc. Natl Acad. Sci. USA 110, 15247–15252 (2013).
Fiil, B. K. et al. OTULIN restricts Met1-linked ubiquitination to control innate immune signaling. Mol. Cell 50, 818–830 (2013).
Rahighi, S. et al. NEMO binding to linear ubiquitin chains is essential for NF-κB activation. Cell 136, 1098–1109 (2009).
Keusekotten, K. et al. OTULIN antagonizes LUBAC signaling by specifically hydrolyzing Met1-linked polyubiquitin. Cell 153, 1312–1326 (2013).
Rivkin, E. et al. The linear ubiquitin-specific deubiquitinase gumby regulates angiogenesis. Nature 498, 318–324 (2013).
Elliott, P. R. et al. Molecular basis and regulation of OTULIN-LUBAC interaction. Mol. Cell 54, 335–348 (2014).
Schaeffer, V. et al. Binding of OTULIN to the PUB domain of HOIP controls NF-kappaB signaling. Mol. Cell 54, 349–361 (2014).
Komander, D. et al. The structure of the CYLD USP domain explains its specificity for Lys63-linked polyubiquitin and reveals a B box module. Mol. Cell 29, 451–464 (2008).
Komander, D. et al. Molecular discrimination of structurally equivalent Lys63-linked and linear polyubiquitin chains. EMBO Rep. 10, 466–473 (2009).
Elliott, P. R. et al. SPATA2 links CYLD to LUBAC, activates CYLD, and controls LUBAC signaling. Mol. Cell 63, 990–1005 (2016).
Kupka, S. et al. SPATA2-mediated binding of CYLD to HOIP enables CYLD recruitment to signaling complexes. Cell Rep. 16, 2271–2280 (2016).
Schlicher, L. et al. SPATA2 promotes CYLD activity and regulates TNF-induced NF-kappaB signaling and cell death. EMBO Rep. 17, 1485–1497 (2016).
Wagner, S. A., Satpathy, S., Beli, P. & Choudhary, C. SPATA2 links CYLD to the TNF-alpha receptor signaling complex and modulates the receptor signaling outcomes. EMBO J. 35, 1868–1884 (2016).
Brummelkamp, T. R., Nijman, S. M., Dirac, A. M. & Bernards, R. Loss of the cylindromatosis tumour suppressor inhibits apoptosis by activating NF-kappaB. Nature 424, 797–801 (2003).
Kovalenko, A. et al. The tumour suppressor CYLD negatively regulates NF-kappaB signalling by deubiquitination. Nature 424, 801–805 (2003).
Trompouki, E. et al. CYLD is a deubiquitinating enzyme that negatively regulates NF-kappaB activation by TNFR family members. Nature 424, 793–796 (2003).
Takiuchi, T. et al. Suppression of LUBAC-mediated linear ubiquitination by a specific interaction between LUBAC and the deubiquitinases CYLD and OTULIN. Genes Cells 19, 254–272 (2014).
Draber, P. et al. LUBAC-recruited CYLD and A20 regulate gene activation and cell death by exerting opposing effects on linear ubiquitin in signaling complexes. Cell Rep. 13, 2258–2272 (2015).
Hrdinka, M. et al. CYLD limits Lys63- and Met1-linked ubiquitin at receptor complexes to regulate innate immune signaling. Cell Rep. 14, 2846–2858 (2016). References 133–135 explore the complex interplay of CYLD, OTULIN and A20 in innate immune signalling.
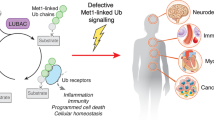
Heger, K. et al. OTULIN limits cell death and inflammation by deubiquitinating LUBAC. Nature 559, 120–124 (2018).
van Wijk, S. J. L. et al. Linear ubiquitination of cytosolic Salmonella Typhimurium activates NF-kappaB and restricts bacterial proliferation. Nat. Microbiol. 2, 17066 (2017).
Bignell, G. R. et al. Identification of the familial cylindromatosis tumour-suppressor gene. Nat. Genet. 25, 160–165 (2000).
Zhou, Q. et al. Biallelic hypomorphic mutations in a linear deubiquitinase define otulipenia, an early-onset autoinflammatory disease. Proc. Natl Acad. Sci. USA 113, 10127–10132 (2016).
Massoumi, R., Chmielarska, K., Hennecke, K., Pfeifer, A. & Fassler, R. Cyld inhibits tumor cell proliferation by blocking Bcl-3-dependent NF-kappaB signaling. Cell 125, 665–677 (2006).
Reiley, W. W. et al. Regulation of T cell development by the deubiquitinating enzyme CYLD. Nat. Immunol. 7, 411–417 (2006).
Zhang, J. et al. Impaired regulation of NF-kappaB and increased susceptibility to colitis-associated tumorigenesis in CYLD-deficient mice. J. Clin. Invest. 116, 3042–3049 (2006).
Jin, W. et al. Deubiquitinating enzyme CYLD regulates the peripheral development and naive phenotype maintenance of B cells. J. Biol. Chem. 282, 15884–15893 (2007).
Reiley, W. W. et al. Deubiquitinating enzyme CYLD negatively regulates the ubiquitin-dependent kinase Tak1 and prevents abnormal T cell responses. J. Exp. Med. 204, 1475–1485 (2007).
Chu, Y. et al. A20 and CYLD do not share significant overlapping functions during B cell development and activation. J. Immunol. 189, 4437–4443 (2012).
Peltzer, N. et al. HOIP deficiency causes embryonic lethality by aberrant TNFR1-mediated endothelial cell death. Cell Rep. 9, 153–165 (2014).
Peltzer, N. et al. LUBAC is essential for embryogenesis by preventing cell death and enabling haematopoiesis. Nature 557, 112–117 (2018).
Morrow, M. E. et al. Active site alanine mutations convert deubiquitinases into high-affinity ubiquitin-binding proteins. EMBO Rep. 19, e45680 (2018).
Boone, D. L. et al. The ubiquitin-modifying enzyme A20 is required for termination of Toll-like receptor responses. Nat. Immunol. 5, 1052–1060 (2004).
Wertz, I. E. et al. De-ubiquitination and ubiquitin ligase domains of A20 downregulate NF-kappaB signalling. Nature 430, 694–699 (2004).
Catrysse, L., Vereecke, L., Beyaert, R. & van Loo, G. A20 in inflammation and autoimmunity. Trends Immunol. 35, 22–31 (2014).
Komander, D. & Barford, D. Structure of the A20 OTU domain and mechanistic insights into deubiquitination. Biochem. J. 409, 77–85 (2008).
Wertz, I. E. et al. Phosphorylation and linear ubiquitin direct A20 inhibition of inflammation. Nature 528, 370–375 (2015).
Ohtake, F., Saeki, Y., Ishido, S., Kanno, J. & Tanaka, K. The K48-K63 branched ubiquitin chain regulates NF-kappaB signaling. Mol. Cell 64, 251–266 (2016).
Lu, T. T. et al. Dimerization and ubiquitin mediated recruitment of A20, a complex deubiquitinating enzyme. Immunity 38, 896–905 (2013).
De, A., Dainichi, T., Rathinam, C. V. & Ghosh, S. The deubiquitinase activity of A20 is dispensable for NF-kappaB signaling. EMBO Rep. 15, 775–783 (2014).
Lee, E. G. et al. Failure to regulate TNF-induced NF-kappaB and cell death responses in A20-deficient mice. Science 289, 2350–2354 (2000).
Zhao, Y. et al. OTUD4 is a phospho-activated K63 deubiquitinase that regulates MyD88-dependent signaling. Mol. Cell 69, 505–516 (2018). This paper demonstrates that a switch in chain linkage specificity, mediated by phosphorylation of OTUD4, unleashes Lys63 chain-directed activity to negatively regulate Toll-like receptor-mediated activation of the NF-κB pathway.
Clague, M. J., Liu, H. & Urbe, S. Governance of endocytic trafficking and signaling by reversible ubiquitylation. Dev. Cell 23, 457–467 (2012).
Huang, F., Kirkpatrick, D., Jiang, X., Gygi, S. & Sorkin, A. Differential regulation of EGF receptor internalization and degradation by multiubiquitination within the kinase domain. Mol. Cell 21, 737–748 (2006).
Huang, F. et al. Lysine 63-linked polyubiquitination is required for EGF receptor degradation. Proc. Natl Acad. Sci. USA 110, 15722–15727 (2013).
Row, P. E. et al. The MIT domain of UBPY constitutes a CHMP binding and endosomal localization signal required for efficient epidermal growth factor receptor degradation. J. Biol. Chem. 282, 30929–30937 (2007).
Crespo-Yanez, X. et al. CHMP1B is a target of USP8/UBPY regulated by ubiquitin during endocytosis. PLOS Genet. 14, e1007456 (2018).
Row, P. E., Prior, I. A., McCullough, J., Clague, M. J. & Urbe, S. The ubiquitin isopeptidase UBPY regulates endosomal ubiquitin dynamics and is essential for receptor down-regulation. J. Biol. Chem. 281, 12618–12624 (2006).
McCullough, J., Clague, M. J. & Urbe, S. AMSH is an endosome-associated ubiquitin isopeptidase. J. Cell Biol. 166, 487–492 (2004).
McDonell, L. M. et al. Mutations in STAMBP, encoding a deubiquitinating enzyme, cause microcephaly-capillary malformation syndrome. Nat. Genet. 45, 556–562 (2013).
Shrestha, R. K. et al. Insights into the mechanism of deubiquitination by JAMM deubiquitinases from cocrystal structures of the enzyme with the substrate and product. Biochemistry 53, 3199–3217 (2014).
Ma, Z. Y. et al. Recurrent gain-of-function USP8 mutations in Cushing’s disease. Cell Res. 25, 306–317 (2015). This paper reports identification of mutations in USP8 from exome sequencing of corticotroph adenomas. Mutations inhibit 14-3-3 protein binding, resulting in gain-of-function increased activity.
Reincke, M. et al. Mutations in the deubiquitinase gene USP8 cause Cushing’s disease. Nat. Genet. 47, 31–38 (2015).
Clague, M. J. & Urbe, S. Endocytosis: the DUB version. Trends Cell Biol. 16, 551–559 (2006).
Lu, K., den Brave, F. & Jentsch, S. Receptor oligomerization guides pathway choice between proteasomal and autophagic degradation. Nat. Cell Biol. 19, 732–739 (2017).
Clague, M. J. & Urbe, S. Ubiquitin: same molecule, different degradation pathways. Cell 143, 682–685 (2010).
Harper, J. W., Ordureau, A. & Heo, J. M. Building and decoding ubiquitin chains for mitophagy. Nat. Rev. Mol. Cell Biol. 19, 93–108 (2018).
Ordureau, A. et al. Quantitative proteomics reveal a feedforward mechanism for mitochondrial PARKIN translocation and ubiquitin chain synthesis. Mol. Cell 56, 360–375 (2014).
Cunningham, C. N. et al. USP30 and parkin homeostatically regulate atypical ubiquitin chains on mitochondria. Nat. Cell Biol. 17, 160–169 (2015).
Durcan, T. M. et al. USP8 regulates mitophagy by removing K6-linked ubiquitin conjugates from parkin. EMBO J. 33, 2473–2491 (2014).
Nakamura, N. & Hirose, S. Regulation of mitochondrial morphology by USP30, a deubiquitinating enzyme present in the mitochondrial outer membrane. Mol. Biol. Cell 19, 1903–1911 (2008).
Bingol, B. et al. The mitochondrial deubiquitinase USP30 opposes parkin-mediated mitophagy. Nature 510, 370–375 (2014). This paper shows that the mitochondrial DUB USP30 can suppress PINK1-mediated and parkin-mediated mitophagy. USP30 depletion corrects Parkinson disease-related phenotypes in PINK1 mutant and parkin mutant fly models.
Liang, J. R. et al. USP30 deubiquitylates mitochondrial Parkin substrates and restricts apoptotic cell death. EMBO Rep. 16, 618–627 (2015).
Wauer, T. et al. Ubiquitin Ser65 phosphorylation affects ubiquitin structure, chain assembly and hydrolysis. EMBO J. 34, 307–325 (2015).
Kane, L. A. et al. PINK1 phosphorylates ubiquitin to activate Parkin E3 ubiquitin ligase activity. J. Cell Biol. 205, 143–153 (2014).
Kazlauskaite, A. et al. Phosphorylation of Parkin at Serine65 is essential for activation: elaboration of a Miro1 substrate-based assay of Parkin E3 ligase activity. Open Biol. 4, 130213 (2014).
Kazlauskaite, A. et al. Parkin is activated by PINK1-dependent phosphorylation of ubiquitin at Ser65. Biochem. J. 460, 127–139 (2014).
Ohtake, F. et al. Ubiquitin acetylation inhibits polyubiquitin chain elongation. EMBO Rep. 16, 192–201 (2014).
Huguenin-Dezot, N. et al. Synthesis of isomeric phosphoubiquitin chains reveals that phosphorylation controls deubiquitinase activity and specificity. Cell Rep. 16, 1180–1193 (2016).
Clague, M. J. & Urbe, S. Integration of cellular ubiquitin and membrane traffic systems: focus on deubiquitylases. FEBS J. 284, 1753–1766 (2017).
Matsumoto, M. L. et al. K11-linked polyubiquitination in cell cycle control revealed by a K11 linkage-specific antibody. Mol. Cell. 39, 477–484 (2010).
Garnett, M. J. et al. UBE2S elongates ubiquitin chains on APC/C substrates to promote mitotic exit. Nat. Cell Biol. 11, 1363–1369 (2009).
Wickliffe, K. E., Lorenz, S., Wemmer, D. E., Kuriyan, J. & Rape, M. The mechanism of linkage-specific ubiquitin chain elongation by a single-subunit E2. Cell 144, 769–781 (2011).
Williamson, A. et al. Regulation of ubiquitin chain initiation to control the timing of substrate degradation. Mol. Cell 42, 744–757 (2011).
Brown, N. G. et al. Dual RING E3 architectures regulate multiubiquitination and ubiquitin chain elongation by APC/C. Cell 165, 1440–1453 (2016).
Yau, R. G. et al. Assembly and function of heterotypic ubiquitin chains in cell-cycle and protein quality control. Cell 171, 918–933 (2017).
Bremm, A., Freund, S. M. & Komander, D. Lys11-linked ubiquitin chains adopt compact conformations and are preferentially hydrolyzed by the deubiquitinase Cezanne. Nat. Struct. Mol. Biol. 17, 939–947 (2010).
Bonacci, T. et al. Cezanne/OTUD7B is a cell cycle-regulated deubiquitinase that antagonizes the degradation of APC/C substrates. EMBO J. 37, e98701 (2018).
Silva, G. O. et al. Cross-species DNA copy number analyses identifies multiple 1q21-q23 subtype-specific driver genes for breast cancer. Breast Cancer Res. Treat. 152, 347–356 (2015).
Ryu, K. Y., Baker, R. T. & Kopito, R. R. Ubiquitin-specific protease 2 as a tool for quantification of total ubiquitin levels in biological specimens. Anal. Biochem. 353, 153–155 (2006).
Hospenthal, M. K., Mevissen, T. E. T. & Komander, D. Deubiquitinase-based analysis of ubiquitin chain architecture using Ubiquitin Chain Restriction (UbiCRest). Nat. Protoc. 10, 349–361 (2015). This study introduces the restriction analysis of ubiquitin chain architecture using a panel of DUBs with defined specificities.
Harrigan, J. A., Jacq, X., Martin, N. M. & Jackson, S. P. Deubiquitylating enzymes and drug discovery: emerging opportunities. Nat. Rev. Drug Discov. 17, 57–78 (2018).
Schwickart, M. et al. Deubiquitinase USP9X stabilizes MCL1 and promotes tumour cell survival. Nature 463, 103–107 (2010).
Schulein-Volk, C. et al. Dual regulation of Fbw7 function and oncogenic transformation by Usp28. Cell Rep. 9, 1099–1109 (2014).
Tavana, O. et al. HAUSP deubiquitinates and stabilizes N-Myc in neuroblastoma. Nat. Med. 22, 1180–1186 (2016).
Tavana, O., Sun, H. & Gu, W. Targeting HAUSP in both p53 wildtype and p53-mutant tumors. Cell Cycle 17, 823–828 (2018).
Zhang, S. et al. Deubiquitinase USP13 dictates MCL1 stability and sensitivity to BH3 mimetic inhibitors. Nat. Commun. 9, 215 (2018).
Brooks, C. L. & Gu, W. p53 ubiquitination: Mdm2 and beyond. Mol. Cell 21, 307–315 (2006).
Kategaya, L. et al. USP7 small-molecule inhibitors interfere with ubiquitin binding. Nature 550, 534–538 (2017).
Lamberto, I. et al. Structure-guided development of a potent and selective non-covalent active-site inhibitor of USP7. Cell Chem. Biol. 24, 1490–1500 (2017).
Turnbull, A. P. et al. Molecular basis of USP7 inhibition by selective small-molecule inhibitors. Nature 550, 481–486 (2017).
Gavory, G. et al. Discovery and characterization of highly potent and selective allosteric USP7 inhibitors. Nat. Chem. Biol. 14, 118–125 (2018). References 205–208 introduce highly specific USP7 inhibitors with accompanying structural analyses and descriptions of biological consequences, such as elevation of p53 levels.
Faesen, A. C. et al. Mechanism of USP7/HAUSP activation by its C-terminal ubiquitin-like domain and allosteric regulation by GMP-synthetase. Mol. Cell 44, 147–159 (2011).
Chauhan, D. et al. A small molecule inhibitor of ubiquitin-specific protease-7 induces apoptosis in multiple myeloma cells and overcomes bortezomib resistance. Cancer Cell 22, 345–358 (2012).
Song, M. S. et al. The deubiquitinylation and localization of PTEN are regulated by a HAUSP-PML network. Nature 455, 813–817 (2008).
Faustrup, H., Bekker-Jensen, S., Bartek, J., Lukas, J. & Mailand, N. USP7 counteracts SCFbetaTrCP- but not APCCdh1-mediated proteolysis of Claspin. J. Cell Biol. 184, 13–19 (2009).
van Loosdregt, J. et al. Stabilization of the transcription factor Foxp3 by the deubiquitinase USP7 increases Treg-cell-suppressive capacity. Immunity 39, 259–271 (2013).
Li, J. et al. Epidithiodiketopiperazines inhibit protein degradation by targeting proteasome deubiquitinase Rpn11. Cell Chem. Biol. 25, 1350–1358 (2018).
Lee, B. H. et al. Enhancement of proteasome activity by a small-molecule inhibitor of USP14. Nature 467, 179–184 (2010).
Homma, T. et al. Ubiquitin-specific protease 14 modulates degradation of cellular prion protein. Sci. Rep. 5, 11028 (2015).
McKinnon, C. et al. Prion-mediated neurodegeneration is associated with early impairment of the ubiquitin-proteasome system. Acta Neuropathol. 131, 411–425 (2016).
Wang, Y. et al. Small molecule inhibitors reveal allosteric regulation of USP14 via steric blockade. Cell Res. 28, 1186–1194 (2018).
McCullough, J. et al. Activation of the endosome-associated ubiquitin isopeptidase AMSH by STAM, a component of the multivesicular body-sorting machinery. Curr. Biol. 16, 160–165 (2006).
Cooper, E. M., Boeke, J. D. & Cohen, R. E. Specificity of the BRISC deubiquitinating enzyme is not due to selective binding to Lys63-linked polyubiquitin. J. Biol. Chem. 285, 10344–10352 (2010).
Virdee, S., Ye, Y., Nguyen, D. P., Komander, D. & Chin, J. W. Engineered diubiquitin synthesis reveals Lys-29-isopeptide specificity of an OTU deubiquitinase. Nat. Chem. Biol. 6, 750–756 (2010).
Pai, M. T. et al. Solution structure of the Ubp-M BUZ domain, a highly specific protein module that recognizes the C-terminal tail of free ubiquitin. J. Mol. Biol. 370, 290–302 (2007).
Nicassio, F. et al. Human USP3 is a chromatin modifier required for S phase progression and genome stability. Curr. Biol. 17, 1972–1977 (2007).
Acknowledgements
The authors thank J. Jardine for help with the preparation of original figures and K. Hofmann, D. Rigden, R. Damgaard and J. Morris for advice on sections of the manuscript.
Reviewer information
Nature Reviews Molecular Cell Biology thanks N. Mailand and other anonymous reviewer(s) for their contribution to the peer review of this work.
Author information
Authors and Affiliations
Contributions
M.J.C. wrote the manuscript with input from D.K. and S.U. S.U. prepared original figures. All authors read and discussed the manuscript and responded to reviewers.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no conflicts of interest.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Glossary
- SUMO
-
A family of ubiquitin-like proteins (SUMO1, SUMO2 and SUMO3) that can be attached to lysines of target proteins in a similar way to ubiquitin.
- NEDD8
-
A ubiquitin-like protein that is conjugated to and activates a large family of ubiquitin E3 ligases: cullin–RING E3 ligases.
- ISG15
-
A ubiquitin-like protein that is composed of two ubiquitin-like domains and is induced in response to interferon. ISG15 can be conjugated to proteins but also has activity in its unconjugated or even its secreted form.
- Pseudoenzymes
-
Catalytically deficient variants of enzymes that exist in all major enzyme families.
- COP9 signalosome
-
An eight-subunit protein complex that regulates protein ubiquitylation and turnover in various developmental and physiological contexts. Extensively characterized in plants but fundamental to all eukaryotes, this complex post-translationally modifies the cullin subunit of E3 ubiquitin ligases by cleaving off NEDD8.
- Housekeeping functions
-
Basic or fundamental functions common to most cell types that maintain the broad cellular infrastructure.
- Cullins
-
A family of scaffold proteins that constitute the backbone of a large superfamily of ubiquitin E3 ligases (cullin RING ligases (CRLs)).
- Stress granules
-
Dense cytosolic assemblies or aggregates of ribosomal RNA and protein that accumulate in response to stress.
- AAA-ATPases
-
A large superfamily of ATPases that regulate diverse processes in cells including disaggregation of proteins, protein degradation and membrane traffic.
- APC/C
-
A large multiprotein ubiquitin E3 ligase complex that orchestrates cell cycle progression by promoting the proteasomal degradation of key cell cycle regulators.
- Chromatin remodelling
-
Dynamic changes in the chromatin architecture that regulate access to DNA.
- U4/U6.U5 triple small nuclear ribonucleoprotein
-
(U4/U6.U5 tri-snRNP). A major building block of the spliceosome comprising U4, U6 and U5 small nuclear RNAs (snRNAs) (with U4 and U6 strongly base paired) and >30 proteins.
- Pulse–chase studies
-
Experiments that follow the fate of newly synthesized proteins over time by labelling proteins with radioactive isotope-containing or stable isotope-containing amino acids for a short time (pulse) followed by a chase with unlabelled amino acids.
- Fanconi anaemia DNA crosslink repair
-
A DNA damage repair pathway that resolves DNA interstrand crosslinks and is executed by >20 proteins, for which loss of function is associated with Fanconi anaemia, a recessive disorder characterized by chromosomal instability and hypersensitivity to agents that induce DNA crosslinks.
- Translesion synthesis repair
-
An error-prone DNA damage repair process that allows DNA replication to proceed past lesions such as thymidine dimers or abasic sites using specialized DNA polymerases.
- γ-H2AX
-
A histone H2AX variant that is phosphorylated on Ser139 and accumulates rapidly in response to double-stranded DNA damage.
- Linker histone H1
-
A histone that binds to the linker DNA in-between nucleosomes, which are themselves composed of an octamer of histones H2A, H2B, H3 and H4.
- Non-homologous end joining
-
(NHEJ). A DNA damage repair pathway that does not rely on the presence of a homologous template, in contrast to homology-directed repair.
- Homology-directed repair
-
(HDR). A mechanism of double-strand DNA damage repair that relies on a homologous template and thus is restricted to the G2 and S phases of the cell cycle.
- Replication stress
-
Stress occurring during genome replication, a major cause of genome instability.
- Micronuclei
-
Extranuclear bodies containing chromosomes or fragments thereof that escaped incorporation into the nucleus during cell division.
- NF-κB signalling pathway
-
An innate immune signalling pathway that results in the activation of the transcription factor nuclear factor-κB (NF-κB).
- Pattern recognition receptors
-
Receptors including Toll-like receptors and NOD that recognize conserved molecular structures (pathogen-associated molecular patterns and damage-associated molecular patterns) that are found in pathogens (bacteria, viruses, fungi and parasites).
- Endosomal sorting complex required for transport
-
(ESCRT). A multimeric protein complex that was first identified biochemically in yeast. One function of the ESCRT machinery is to control the sorting of endosomal cargo proteins into internal vesicles of multivesicular bodies.
- Microcephaly capillary malformation syndrome
-
(MIC-CAP). An inherited (congenital) disorder characterized by an abnormally small head and aberrant capillaries in the skin.
- Cushing disease
-
A collection of symptoms caused by prolonged exposure to high levels of cortisol in the blood, most commonly caused by a benign tumour of the pituitary gland (resulting in increased levels of adrenocorticotropic hormone release and stimulation of cortisol production in the adrenal gland).
- Securin
-
A key mitotic substrate of APC/C that needs to be degraded to allow the segregation of sister chromatids during anaphase.
Rights and permissions
About this article
Cite this article
Clague, M.J., Urbé, S. & Komander, D. Breaking the chains: deubiquitylating enzyme specificity begets function. Nat Rev Mol Cell Biol 20, 338–352 (2019). https://doi.org/10.1038/s41580-019-0099-1
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41580-019-0099-1
This article is cited by
-
Stabilization of Pin1 by USP34 promotes Ubc9 isomerization and protein sumoylation in glioma stem cells
Nature Communications (2024)
-
Proteome-scale discovery of protein degradation and stabilization effectors
Nature (2024)
-
VDR and deubiquitination control neuronal oxidative stress and microglial inflammation in Parkinson’s disease
Cell Death Discovery (2024)
-
Emerging roles of deubiquitinating enzymes in actin cytoskeleton and tumor metastasis
Cellular Oncology (2024)
-
OTULIN Haploinsufficiency-Related Fasciitis and Skin Necrosis Treated by TNF Inhibition
Journal of Clinical Immunology (2024)