Abstract
The oncoprotein Smoothened (SMO), a G-protein-coupled receptor (GPCR) of the Frizzled-class (class-F), transduces the Hedgehog signal from the tumour suppressor Patched-1 (PTCH1) to the glioma-associated-oncogene (GLI) transcription factors, which activates the Hedgehog signalling pathway1,2. It has remained unknown how PTCH1 modulates SMO, how SMO is stimulated to form a complex with heterotrimeric G proteins and whether G-protein coupling contributes to the activation of GLI proteins3. Here we show that 24,25-epoxycholesterol, which we identify as an endogenous ligand of PTCH1, can stimulate Hedgehog signalling in cells and can trigger G-protein signalling via human SMO in vitro. We present a cryo-electron microscopy structure of human SMO bound to 24(S),25-epoxycholesterol and coupled to a heterotrimeric Gi protein. The structure reveals a ligand-binding site for 24(S),25-epoxycholesterol in the 7-transmembrane region, as well as a Gi-coupled activation mechanism of human SMO. Notably, the Gi protein presents a different arrangement from that of class-A GPCR–Gi complexes. Our work provides molecular insights into Hedgehog signal transduction and the activation of a class-F GPCR.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
Data availability
The data that support the findings of this study are available from the corresponding author upon request. The 3D cryo-EM density map has been deposited in the Electron Microscopy Data Bank under the accession number EMD-20190. Atomic coordinates for the atomic model have been deposited in the Protein Data Bank under the accession number 6OT0.
References
Jiang, J. & Hui, C. C. Hedgehog signaling in development and cancer. Dev. Cell 15, 801–812 (2008).
Petrov, K., Wierbowski, B. M. & Salic, A. Sending and receiving Hedgehog signals. Annu. Rev. Cell Dev. Biol. 33, 145–168 (2017).
Arensdorf, A. M., Marada, S. & Ogden, S. K. Smoothened regulation: a tale of two signals. Trends Pharmacol. Sci. 37, 62–72 (2016).
Taipale, J., Cooper, M. K., Maiti, T. & Beachy, P. A. Patched acts catalytically to suppress the activity of Smoothened. Nature 418, 892–896 (2002).
Qi, X., Schmiege, P., Coutavas, E., Wang, J. & Li, X. Structures of human Patched and its complex with native palmitoylated sonic hedgehog. Nature 560, 128–132 (2018).
Qi, X., Schmiege, P., Coutavas, E. & Li, X. Two Patched molecules engage distinct sites on Hedgehog yielding a signaling-competent complex. Science 362, eaas8843 (2018).
Zhang, Y. et al. Structural basis for cholesterol transport-like activity of the Hedgehog receptor Patched. Cell 175, 1352–1364.e1314 (2018).
Pak, E. & Segal, R. A. Hedgehog signal transduction: key players, oncogenic drivers, and cancer therapy. Dev. Cell 38, 333–344 (2016).
Dunaeva, M., Michelson, P., Kogerman, P. & Toftgard, R. Characterization of the physical interaction of Gli proteins with SUFU proteins. J. Biol. Chem. 278, 5116–5122 (2003).
Riobo, N. A., Saucy, B., Dilizio, C. & Manning, D. R. Activation of heterotrimeric G proteins by Smoothened. Proc. Natl Acad. Sci. USA 103, 12607–12612 (2006).
Ogden, S. K. et al. G protein Gαi functions immediately downstream of Smoothened in Hedgehog signalling. Nature 456, 967–970 (2008).
Byrne, E. F. X. et al. Structural basis of Smoothened regulation by its extracellular domains. Nature 535, 517–522 (2016).
Huang, P. et al. Cellular cholesterol directly activates Smoothened in Hedgehog signaling. Cell 166, 1176–1187 (2016).
Huang, P. et al. Structural basis of Smoothened activation in Hedgehog signaling. Cell 175, 295–297 (2018).
Wang, C. et al. Structure of the human smoothened receptor bound to an antitumour agent. Nature 497, 338–343 (2013).
Wang, C. et al. Structural basis for Smoothened receptor modulation and chemoresistance to anticancer drugs. Nat. Commun. 5, 4355 (2014).
Huang, H. C. & Klein, P. S. The Frizzled family: receptors for multiple signal transduction pathways. Genome Biol. 5, 234 (2004).
Sharpe, H. J., Wang, W., Hannoush, R. N. & de Sauvage, F. J. Regulation of the oncoprotein Smoothened by small molecules. Nat. Chem. Biol. 11, 246–255 (2015).
Raleigh, D. R. et al. Cilia-associated oxysterols activate Smoothened. Mol. Cell 72, 316–327.e315 (2018).
Chen, J. K., Taipale, J., Cooper, M. K. & Beachy, P. A. Inhibition of Hedgehog signaling by direct binding of cyclopamine to Smoothened. Genes Dev. 16, 2743–2748 (2002).
Nachtergaele, S. et al. Structure and function of the Smoothened extracellular domain in vertebrate Hedgehog signaling. eLife 2, e01340 (2013).
Kang, Y. et al. Cryo-EM structure of human rhodopsin bound to an inhibitory G protein. Nature 558, 553–558 (2018).
Myers, B. R. et al. Hedgehog pathway modulation by multiple lipid binding sites on the smoothened effector of signal response. Dev. Cell 26, 346–357 (2013).
Weierstall, U. et al. Lipidic cubic phase injector facilitates membrane protein serial femtosecond crystallography. Nat. Commun. 5, 3309 (2014).
Draper-Joyce, C. J. et al. Structure of the adenosine-bound human adenosine A1 receptor-Gi complex. Nature 558, 559–563 (2018).
García-Nafría, J., Nehmé, R., Edwards, P. C. & Tate, C. G. Cryo-EM structure of the serotonin 5-HT1B receptor coupled to heterotrimeric Go. Nature 558, 620–623 (2018).
Koehl, A. et al. Structure of the µ-opioid receptor-Gi protein complex. Nature 558, 547–552 (2018).
Krishna Kumar, K. et al. Structure of a signaling cannabinoid receptor 1-G protein complex. Cell 176, 448–458.e412 (2019).
Wright, S. C. et al. A conserved molecular switch in Class F receptors regulates receptor activation and pathway selection. Nat. Commun. 10, 667 (2019).
Huang, W. et al. Structural insights into µ-opioid receptor activation. Nature 524, 315–321 (2015).
Yang, S. et al. Crystal structure of the Frizzled 4 receptor in a ligand-free state. Nature 560, 666–670 (2018).
Corcoran, R. B. & Scott, M. P. Oxysterols stimulate Sonic hedgehog signal transduction and proliferation of medulloblastoma cells. Proc. Natl Acad. Sci. USA 103, 8408–8413 (2006).
Chen, W. et al. Activity-dependent internalization of smoothened mediated by β-arrestin 2 and GRK2. Science 306, 2257–2260 (2004).
McDonald, J. G., Smith, D. D., Stiles, A. R. & Russell, D. W. A comprehensive method for extraction and quantitative analysis of sterols and secosteroids from human plasma. J. Lipid Res. 53, 1399–1409 (2012).
Rasmussen, S. G. et al. Crystal structure of the β2 adrenergic receptor–Gs protein complex. Nature 477, 549–555 (2011).
Zheng, S. Q. et al. MotionCor2: anisotropic correction of beam-induced motion for improved cryo-electron microscopy. Nat. Methods 14, 331–332 (2017).
Rohou, A. & Grigorieff, N. CTFFIND4: Fast and accurate defocus estimation from electron micrographs. J. Struct. Biol. 192, 216–221 (2015).
Scheres, S. H. RELION: implementation of a Bayesian approach to cryo-EM structure determination. J. Struct. Biol. 180, 519–530 (2012).
Grigorieff, N. Frealign: an exploratory tool for single-particle cryo-EM. Methods Enzymol. 579, 191–226 (2016).
Emsley, P. & Cowtan, K. Coot: model-building tools for molecular graphics. Acta Crystallogr. D 60, 2126–2132 (2004).
Adams, P. D. et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D 66, 213–221 (2010).
Murshudov, G. N., Vagin, A. A. & Dodson, E. J. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr. D 53, 240–255 (1997).
Brown, A. et al. Tools for macromolecular model building and refinement into electron cryo-microscopy reconstructions. Acta Crystallogr. D 71, 136–153 (2015).
Ten Eyck, L. F. Efficient structure-factor calculation for large molecules by the fast fourier transform. Acta Crystallogr. A 33, 486–492 (1977).
Wang, Z. et al. An atomic model of brome mosaic virus using direct electron detection and real-space optimization. Nat. Commun. 5, 4808 (2014).
Heymann, J. B. & Belnap, D. M. Bsoft: image processing and molecular modeling for electron microscopy. J. Struct. Biol. 157, 3–18 (2007).
Chen, V. B. et al. MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr. D 66, 12–21 (2010).
Pettersen, E. F. et al. UCSF Chimera—a visualization system for exploratory research and analysis. J. Comput. Chem. 25, 1605–1612 (2004).
Acknowledgements
The data were collected at the UT Southwestern Medical Center Cryo-EM Facility (funded in part by the CPRIT Core Facility Support Award RP170644); we thank D. Stoddard for assistance in data collection; E. Xu for sharing the materials for making Fab-G50; L. Beatty, L. Friedberg, A. Hassan and P. Schmiege for technical help; M. Brown, E. Debler, J. Goldstein and J. Jiang for discussion during manuscript preparation; B. Chen and J. Kim for Smo−/− mouse embryonic fibroblasts. This work was supported by the Endowed Scholars Program in Medical Science of UT Southwestern Medical Center (to X.L.), O’Donnell Junior Faculty Funds (to X.L.), Welch Foundation (I-1957) (to X.L.), NIH grant P01 HL020948 (to J.M. and X.L.) and NIH grant 1R35GM128641 (to C.Z.). X.L. is a Damon Runyon-Rachleff Innovator supported by the Damon Runyon Cancer Research Foundation (DRR-53-19) and a Rita C. and William P. Clements Jr. Scholar in Biomedical Research at UT Southwestern Medical Center.
Author information
Authors and Affiliations
Contributions
X.Q. and X.L. conceived the project and designed the research with H.L and C.Z. B.T. and J.M. performed the sterol analysis by mass spectrometry. X.Q. purified the protein for mass spectrometry analysis, and H.L. purified the protein for the cryo-EM study. X.Q. carried out cryo-EM work, built the model and refined the structure. X.Q. and H.L. performed the functional characterization. All the authors analysed the data. X.Q., H.L., C.Z. and X.L. contributed to manuscript preparation. X.L. wrote the manuscript. Z.C. and X.L. supervised the project.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data figures and tables
Extended Data Fig. 1 Assembly of hSMO–Gi–Fab complex.
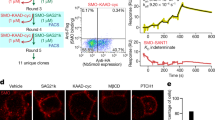
a, GTPγS causes the dissociation of the 24(S),25-EC-mediated hSMO–Gi complex. b, Size-exclusion chromatogram and SDS–PAGE gel of the purified hSMO–Gi–Fab complex. Molecular standards are indicated on left side of the gel.
Extended Data Fig. 2 Data processing.
a, A representative electron micrograph at −2.0 μm defocus. b, The data-processing workflow for the complex with the full map. The cryo-EM 2D classification from RELION is shown. The subtracted parts are indicated by dashed circles. c, The data-processing workflow for the complex with the subtracted map. Class 3 of the full map and class 4 of the subtracted map were used for the final refinement; class 4 of the full map and class 1 of the subtracted map did not show sufficient structural features in the final refinement. Masks used for the refinement are shown. The cryo-EM map after FREALIGN refinement sharpened using BFACTOR with a resolution limit of 4 Å or 3.9 Å, and a B-factor value of −100 Å2. Each subunit is coloured in a different colour.
Extended Data Fig. 3 Assessment of model quality.
a, FSC curve of the structure without the CRD and half of the Fab, with FSC as a function of resolution, using FREALIGN output. b, The FSC curves calculated between the refined structure and the half map used for refinement (blue), the other half map (red) and the full map (black). c, Density maps of structure, coloured by local-resolution estimation using Blocres.
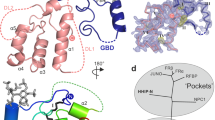
Extended Data Fig. 4 Cryo-EM map of structural elements in the complex.
a, The major helices of hSMO. b, The major structural elements of Gi protein. Electron microscopy density map and model of the complex are shown in mesh and cartoon. c, The putative ligand.
Extended Data Fig. 5 Comparison of the maps in the ligand-binding pocket of hSMO.
a, The extra density within the transmembrane-domain ligand-binding pocket in the hSMO crystal structure (PDB 5L7D). The density is shown in green at the 3σ level, and is indicated by an arrow. b, The density of the ligand in the Gi–hSMO complex. The density is shown in purple mesh at the 5σ level at 3.9 Å, and is indicated by an arrow.
Extended Data Fig. 6 Comparison of the binding sites of different SMO ligands.
a, SAG1.5-bound hSMO (PDB 4QIN). b, Cyclopamine-bound hSMO (PDB 4O9R). c, Vismodegib-bound hSMO (PDB 5L7I). d, SANT1-bound hSMO (PDB 4N4W). e, LY2940680-bound hSMO (PDB 4JKV). In a–e, structures of hSMO with different ligands are viewed from the side of the membrane. f, Superimposition of the ligands that bind the pocket in the transmembrane domain of hSMO.
Extended Data Fig. 7 Comparisons of the TM6 and cytosolic sites of hSMO and μOR.
a, Structural comparison of TM6 of hSMO, μOR and GLP-1R in the inactive and G-protein-bound states. Left, hSMO, inactive SMO in pink (PDB 5L7D). Middle, μOR, inactive μOR in light orange (PDB 4DKL), Gi–μOR in light cyan (PDB 6DDE). Right, GLP-1R, inactive GLP-1R in red (PDB 5VEW), Gs–GLP-1R (PDB 6B3J) in dark blue. b, Electrostatic surface representations of the cytosolic side of SMO and μOR complex with Gαi–α5.
Extended Data Fig. 8 The structures of Gi-bound class-A GPCRs.
a, Rhodopsin–Gi complex (PDB 6CMO). b, A1R–Gi complex (PDB 6D9H). c, μOR–Gi complex (PDB 6DDE). d, CB1–Gi complex (PDB 6N4B).
Extended Data Fig. 9 Comparison of the Gi-coupled hSMO, inactive hSMO and apo human FZD4.
The Gi-coupled hSMO is in blue, the inactive hSMO is in pink (PDB 5L7D) and apo human FZD4 is in yellow (PDB 6BD4). Structures are viewed from the side of the membrane.
Supplementary information
Source data
Rights and permissions
About this article
Cite this article
Qi, X., Liu, H., Thompson, B. et al. Cryo-EM structure of oxysterol-bound human Smoothened coupled to a heterotrimeric Gi. Nature 571, 279–283 (2019). https://doi.org/10.1038/s41586-019-1286-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41586-019-1286-0
This article is cited by
-
A framework for Frizzled-G protein coupling and implications to the PCP signaling pathways
Cell Discovery (2024)
-
Structure, function and drug discovery of GPCR signaling
Molecular Biomedicine (2023)
-
Cellular and molecular mechanisms of Hedgehog signalling
Nature Reviews Molecular Cell Biology (2023)
-
Structural insights into ligand recognition and activation of the medium-chain fatty acid-sensing receptor GPR84
Nature Communications (2023)
-
Revealing the secret behind Smo cholesterylation
Cell Research (2022)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.