-
PDF
- Split View
-
Views
-
Cite
Cite
Karthika Divakaran, Ronald N. Hines, D Gail McCarver, Human Hepatic UGT2B15 Developmental Expression, Toxicological Sciences, Volume 141, Issue 1, September 2014, Pages 292–299, https://doi.org/10.1093/toxsci/kfu126
Close - Share Icon Share
Abstract
Human hepatic UGT2B15 developmental expression changes may alter the metabolism of important drugs and toxicants such as bisphenol A (BPA). Previously, UGT2B15 ontogeny knowledge consisted of transcript data, a dubious surrogate for protein expression. Herein, UGT2B15 protein content was determined in human hepatic microsomes (n = 236, 8 weeks gestation to 18 years). The impact of a common, functional single nucleotide polymorphism (g.253G>T), present in UGT2B15*2 and *5 alleles, was also tested. UGT2B15 expression began during late fetal life, at about 18% of mature values (medians = 48, 267 pmoles/mg of microsomal protein, respectively; p < 0.001). UGT2B15 neonatal (n = 39) and late fetal (≥28 weeks, n = 10) content was similar, but lower than that of infants between 3 and 15 weeks age (n = 46; medians = 38, 48, 404 pmoles/mg microsomal protein, respectively; p < 0.001). Values for the latter group were higher compared with the remaining age group (15 weeks to 18 years; n = 82, p < 0.001). UGT2B15 expression varied 31-fold across the entire sample, and within groups, ranged from 4- to 27-fold. Among postnatal samples, age group, the presence of g.253T and male gender were each significantly associated with greater UGT2B15 expression (p < 0.001, <0.01, and <0.05, respectively; stepwise linear regression). In summary, hepatic UGT2B15 protein onset begins in late gestation; however, the greatest rate of change occurs during the first few weeks after birth. We speculate that the fetus and neonate may have lower clearance of some UGT2B15 substrates, such as BPA, compared with older individuals.
UGT2B15 is a member of the uridine diphosphate-glucuronosyltransferase superfamily of endoplasmic reticulum bound membrane proteins responsible for catalyzing the glucuronidation of a large number of endogenous compounds, environmental chemicals, and therapeutic drugs (Court et al., 2002; Nishiyama et al., 2002; Patel et al., 1995). Recently, the enzyme has been of increased interest because it arguably is critical in bisphenol A (BPA) detoxification (Hanioka et al., 2008). Similar to other UGTs, UGT2B15 transfers glucuronic acid from its cofactor, uridine 5`-diphosphoglucuronic acid, to lipophilic substrates converting them into hydrophilic glucuronides and thereby facilitating their excretion from the body (Miners and MacKenzie, 1991). UGT2B15 shares 78% sequence identity with other UGT2B family members and exhibits characteristic (oxazepam, lorazepam), but overlapping substrate (acetaminophen) and inhibitor (valproic acid) specificities (reviewed in Kiang et al., 2005; Stingl et al., 2013; Tukey and Strassburg, 2000).
Of 10 hepatic UGTs, UGT2B15 expression is among the highest in the adult (Fallon et al., 2013; Ohno and Nakajin, 2009). Thus, based on its abundance, UGT2B15 would be expected to play a substantive metabolic role for any compound with substantial enzyme affinity. For other UGTs, delayed expression onset during development results in impaired metabolism and parent drug toxicity. For example, the lack of mature UGT2B7 expression, which does not occur until about two to three months of age, is the mechanism underlying chloramphenicol-induced gray baby syndrome (Anderson et al., 1997; Weiss et al., 1960). Complete UGT protein expression profiles have not been performed; however, although UGT expression differs with age, each UGT may exhibit a unique maturation profile (de Wildt et al., 1999). For UGT2B15, the developmental data are limited to hepatic mRNA expression levels in two fetal, 16 pediatric and 12 adult liver samples that demonstrate an absence at 20 weeks gestation, but adult transcript levels by seven months of age (Strassburg et al., 2002). Transcript levels do not directly translate to protein levels because of post-transcriptional processing, translational efficiency, and post-translational modifications, each of which may alter protein levels. Thus, quantitation of human UGT2B15 protein levels is needed to provide a more accurate definition of the age-related abundance of this important human hepatic protein.
UGT2B15 genetic differences have been shown to impact UGT2B15-mediated glucuronidation ability for some substrates. The highly prevalent non-synonymous single nucleotide polymorphism (SNP) present in approximately 50% of Caucasians, g.253G>T (D85Y; rs 1902023), is a significant determinant of UGT2B15 interindividual variation, measured both as in vitro metabolic activity (Court et al., 2004; Hanioka et al., 2011) and by UGT2B15-mediated in vivo lorazepam and oxazepam metabolic ability (Chung et al., 2005; He et al., 2009). UGT2B15*2, containing the g.253G>T SNP, is not only associated with decreased enzyme activity (Court et al., 2004) but also decreases UGT2B15 protein levels (Court et al., 2002; Levesque et al., 1997). Both the UGT2B15*2 and UGT2B15*5 alleles contain the g.253G>T SNP. However, the UGT2B15*5 allele also includes a non-synonymous SNP, g.23490A>C (rs 4148269), which, although it alters protein sequence (K523T), has not been shown to significantly affect activity (Court et al., 2004). UGT2B15*3 (L86S), UGT2B15*4 (K523T), and UGT2B15*6 (T352I) are among the less common UGT2B15 alleles and have not been associated with significant functional effects (Court et al., 2004). Thus, the base substitution at g.253, present in both UGT2B15*2 and *5, is the variant of greatest interest; however, changes at both g.253 and g.23490 were evaluated in the current study.
The aims of this study were to determine UGT2B15 ontogeny and to determine the impact of functional genetic differences on inter-subject variation in human UGT2B15 protein expression.
MATERIALS AND METHODS
Chemicals and reagents
UGT2B15 supersomes produced from insect cells infected with baculovirus containing recombinant human UGT2B15 cDNA were purchased from BD Biosciences (Woburn, MA). Purified, recombinant UGT2B15 protein was obtained from Origene Technologies (Rockville, MD). Polyclonal mouse antibody raised against full-length human UGT2B15 was obtained from Novus Biologicals (Littleton, CO). Horseradish peroxidase-conjugated sheep anti-mouse IgG, nitrocellulose membranes, and enhanced chemiluminescence kits were purchased from GE Healthcare Bio-Sciences Corp (Piscataway, NJ). Novex Sharp protein molecular weight standard, Platinum Taq DNA Polymerase High Fidelity, and ABI PRISM SNaPshot Multiplex Kits were purchased from Life Technologies (Grand Island, NY). ExoSap-It and shrimp alkaline phosphatase were purchased from US Biological (Cleveland, OH). All other reagents were obtained from commercial sources at the highest grade available.
Human liver microsomes
Liver tissue samples (N = 236), ranging in age from eight weeks gestation to 18 years, were obtained from the Brain and Tissue Bank of the University of Maryland, Baltimore and the Central Laboratory for Human Embryology at the University of Washington (Seattle, WA) and stored at –80°C until the microsomal fractions were prepared. Samples from individuals likely to have liver disease were excluded. The samples were from 137 males and 82 females (17 were unknown gender). Ethnicity information was available for 200 samples comprising 93 Northern European American, 88 African American, and 19 Hispanic-American subjects. Liver microsomes were prepared by differential centrifugation as described in Koukouritaki et al. (2002).
UGT2B15 immunoquantitation
The amount of UGT2B15 present in the supersomes was quantified using standard curves of recombinant UGT2B15 protein standards (Origene, Rockville, MD). A microsomal sample (1 μg) from each liver, along with aliquots from a single lot of supersomes overexpressing UGT2B15 protein (92–549 fmoles) (BD Biosciences, Woburn, MA), was fractionated by SDS-polyacrylamide gel electrophoresis and transferred to nitrocellulose membranes. The membranes were probed with primary anti-human UGT2B15 antibody (Novus Biologicals, Littleton, CO) followed by secondary horseradish peroxidase-conjugated sheep anti-mouse IgG diluted 1:1000 in tris-buffered saline. The antibody bound UGT2B15 protein was detected by chemiluminescence using an ECL plus Western blotting detection system (GE Healthcare Life Sciences, Piscataway, NJ) per the manufacturer's instructions. The optical densities of the UGT2B15 protein bands were determined using the STORM imaging system (GE Healthcare Life Sciences, Piscataway, NJ). The UGT2B15-specific content in each microsomal sample was calculated by comparison with the UGT2B15 supersome standard curve (minimum acceptable standard curve r2>0.9). The inter assay variability was validated in an analysis of five subjects’ samples, each quantitated in four separate experiments, with both fetal and postnatal samples included among those tested (CV = 3.1–12.7%).
Genotyping
Genomic DNA was isolated from the liver tissue samples using DNAzol reagent or a QIAmp DNA Mini kit as described in Koukouritaki et al. (2002). DNA amplification by polymerase chain reaction (PCR) and single base extension (SBE) methods were used to genotype for the non-synonymous SNPs, g.253G>T and g.23490A>C. PCR reactions (10 μl) contained genomic DNA (50 ng), 3mM MgSO4, 0.2mM each deoxyribonucleotide triphosphate, 0.1μM each primer (g.253G>T: Forward 5`-TGTTGGTCTCCTTGGCATGCAC-3` and Reverse 5`-GATGGCCCCAGGGTTCTAACTG-3`), 0.3μM each primer (g.23490A>C: Forward 5`-ACACTTACTTTCAATCATTTGTGTGACA-3` and Reverse 5`-ATCCAGTAACTCGTCATTTAACATTAGG-3`) (Court et al., 2004), and Platinum Taq DNA Polymerase High Fidelity (1 unit) (Life Technologies, Grand Island, NY). The PCR conditions were: an initial denaturation step at 95°C for 10 min, followed by 40 cycles of denaturation at 95°C for 30 s, annealing at 64°C for 30 s, and elongation at 68°C for 1.5 min, with a final elongation step at 68°C for 15 min. After excess primer and unincorporated deoxynucleotides in the reaction were removed by incubating with ExoSAP-It (United States Biochemical Corp., Cleveland, OH), SBE reactions were performed using single primers (g.253 G>T-Sense: GACTATCCTACATCTTTAACTAAAAAT; g.23490A>C-Anti-sense: CTGACTGACTATAACTAATCTCTTTTCTTCTTCTTTCCT) and the ABI Prism SNaPshot Multiplex Kit (Life Technologies) per the manufacturer's instructions. After a shrimp alkaline phosphatase (United States Biochemical Corp.) clean up step, the SBE reaction products were analyzed by capillary electrophoresis using a 3130xl Genetic Analyzer equipped with GeneMapper software version 4 (Life Technologies). Intact plasmids containing the reference and variant SNP were included as controls for the genotyping reactions.
Statistical analysis
Classification regression tree (CRT) analysis (SPSS Inc., Chicago, IL) was used to divide the samples into age groups. CRT selects optimal strata by limiting the differences within an age group and enhancing differences between age groups. Differences in the UGT2B15 content among the different age groups were confirmed using the Kruskal-Wallis test. Subsequently, stepwise multiple linear regression analysis was used to simultaneously evaluate the putative associations between log-transformed UGT2B15 protein expression and age group, genotype, gender, and ethnicity. An alpha value of ≤0.05 was accepted as significant.
RESULTS
Human Hepatic UGT2B15 Developmental Expression Pattern
The present study determined the human hepatic UGT2B15 developmental expression pattern in 236 microsomal samples (8 weeks gestation to 18 years; n = 71 fetal, 165 postnatal) using SDS-PAGE and Western blotting methods. A single immunoreactive band was observed with a molecular mass near 50 kDa, co-migrating with the corresponding full-length UGT2B15 protein band in the supersomes (see typical blot, Fig. 1).
Western blot analysis of UGT2B15 developmental expression pattern in human liver microsomes: Western blot depicts fractionated microsomal samples from differing age groups representing both fetal and postnatal ages (random order) along with UGT2B15 supersome standards. Electrophoresis and immunodetection using UGT2B15-specific antibody were conducted as described in Materials and Methods.
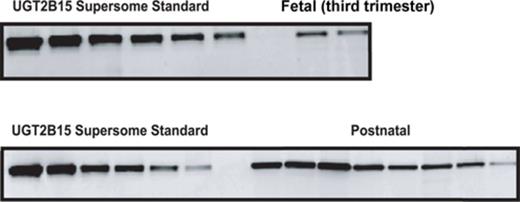
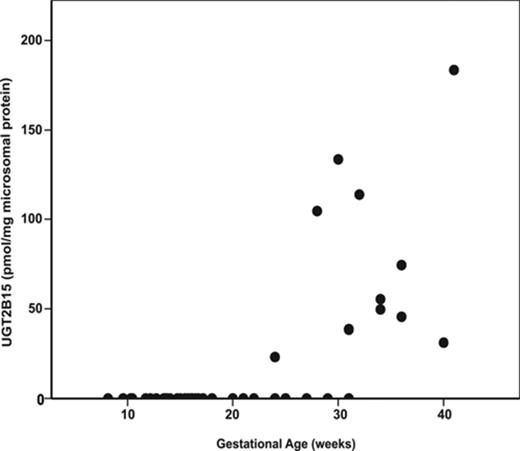
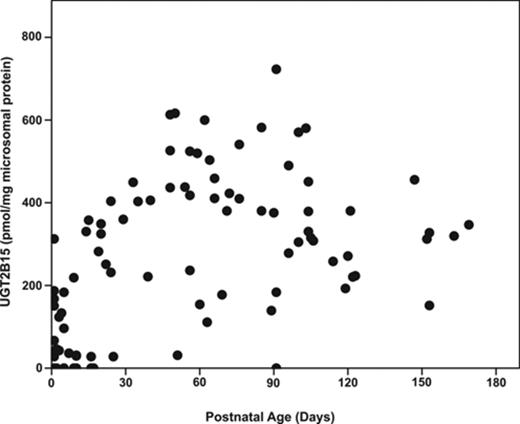
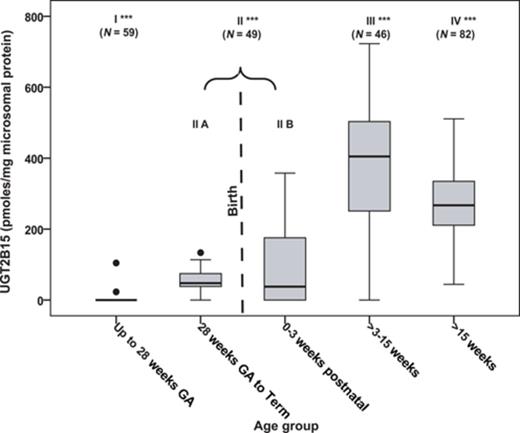
UGT2B15 protein expression was not observed in the first and second trimester liver microsomal samples with the exception of one 24-week gestation fetal liver (Fig. 2). In contrast, the protein was detected in 11 of 13 third trimester fetal livers. UGT2B15 hepatic content in most postnatal samples exceeded 200 pmol/mg microsomal protein, whereas all fetal samples had values less than this (Figs. 2 and 3). UGT2B15 content during the first 90 days of life included many samples with values in the same range as the fetal samples (Figs. 2 and 4). Based on this overlap in values between fetal and early postnatal life, CRT analysis was performed including both fetal and postnatal samples without assuming that birth would, of necessity, alter expression. Four strata were selected using CRT analysis such that UGT2B15 protein expression differed across each comparison: early gestation (≤28 weeks gestational age, n = 59; group I), late gestation and neonatal infancy (28 weeks gestational age to ≤3 weeks postnatal age, n = 49; group II), middle infancy (>3 weeks postnatal age to ≤15 weeks postnatal age, n = 46; group III), and older infancy and childhood (>15 weeks postnatal age to 18 years, n = 82, group IV) (p < 0.001, each comparison; Kruskal Wallace testing) (Fig. 5). Across all four groups, the median microsomal UGT2B15 values varied about sevenfold, with the median among the youngest postnatal bracket, group II, being about 15% of that of the oldest age category, group IV (Fig. 5). Notably, birth did not appear to cause an acute change in UGT2B15 content as the protein expression was similar among samples from individuals between ages 28 weeks gestation and birth (n = 10) and those from birth up to 3 weeks postnatal age (n = 39, p > 0.1; Kruskal Wallace testing) (Fig. 5). The microsomal UGT2B15-specific content was about 10-fold higher in middle infancy (group III), compared with the combined late stage gestation and neonatal infant group (group II) (group III and group II [a+b] median = 405 and 41 pmol/mg microsomal protein, respectively; p < 0.001; Kruskal Wallis testing). UGT2B15 content was higher among samples from individuals older than 15 weeks of age (group IV, median = 267 pmoles/mg microsomal protein; p < 0.001; Kruskal Wallis testing) compared with groups I and II, but lower than that of the middle infant group (ages 3–15 weeks, group III) (p < 0.001; Kruskal Wallis testing).
Human hepatic UGT2B15-specific content as a function of gestational age in 71 fetal hepatic microsomal samples. UGT2B15 content was quantified by Western blotting, densitometry, and comparison with known standards.
Human hepatic UGT2B15-specific protein content as a function of postnatal age in 165 hepatic microsomal samples. UGT2B15 content was quantified by Western blotting, densitometry, and comparison with known standards.
Human hepatic UGT2B15 developmental expression pattern in 97 samples from individuals from birth through the first six postnatal months.
The developmental expression of human hepatic UGT2B15 categorized into four age brackets using CRT analysis. Each of the four major groups differed in UGT2B15 content from every other group. UGT2B15 protein expression was not significantly different among samples between 28 weeks gestational age (GA) and 3 weeks postnatal age. This group, group II, is shown separated into two statistically similar subparts connected by a bracket to visually illustrate the event of birth. The boxes represent the 25th and 75th percentiles, the horizontal bar within the box represents the median, and the vertical bars represent the 5th and 95th percentiles. Outliers (•) were defined as having specific protein content 1.5-fold from the 25th and 75th percentiles. Significant differences between groups were determined by Kruskal-Wallis testing (*** p < 0.001, all between group comparisons).
UGT2B15 Genotype
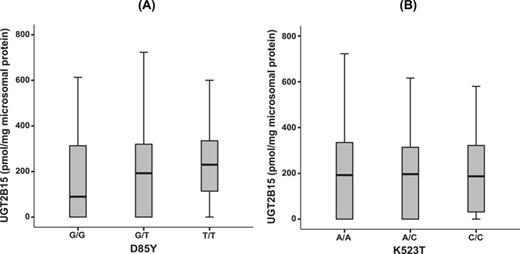
To assess the impact of the common functional UGT2B15 polymorphism, the substitution of a thymidine for guanine at position 253, as well as the impact of the substitution of adenine for cytosine at position 23,490, UGT2B15 genotypes were determined in 225 liver samples (93 Caucasian, 85 African American subjects, 19 Hispanic-American, and 28 of unknown ethnicity). The overall sample genotype frequencies for g.253 GG, GT, and TT were 28%, 47%, and 25%, respectively, and that for g.23490 AA, AC, and CC were 39%, 40%, and 21%, respectively. Among the two ethnic groups with large sample sizes, the genotype frequencies of g.253 GG, GT, and TT were 19%, 52%, and 29% (Caucasians), respectively and 30%, 46%, and 24% (African Americans), respectively. For g.23490 AA, AC, and CC, the frequencies were 24%, 41%, and 35% (Caucasians) and 62%, 34%, and 4% (African Americans), respectively. The genotype distributions were in Hardy-Weinberg equilibrium for the Caucasian and African American populations for UGT2B15 g. 253G>T and g. 23490A>C SNPs. Evaluated independently, the presence of the g.253T variant appeared equivocally associated with increased UGT2B15 expression (Fig. 6A, p = 0.089), whereas g.23490C was not significant (Fig. 6B).
The effect of UGT2B15 genotypes on hepatic UGT2B15-specific content. (A) UGT2B15 protein content in microsomal samples that were homozygous reference (GG), heterozygous (GT), or homozygous variant (TT) for g.253G>T (D85Y). (B) UGT2B15 protein content in microsomal samples that were homozygous reference (AA), heterozygous (AC), or homozygous variant (CC) for g.23490A>C (K523T). The data are represented as box and whisker plots, in which the boxes represent the 25th and 75th percentiles, the horizontal bar within the box represents the median of the data, and the vertical bars represent the 5th and 95th percentiles (N = 225, p = 0.089 and not significant (NS), for D85Y and K523T, respectively; One-way ANOVA).
Factors that May Influence UGT2B15 Developmental Protein Expression
Across the entire dataset, UGT2B15 expression varied 31-fold with 4- to 27-fold interindividual variability within each age group. For the entire sample, age category remained strongly associated with UGT2B15 content when the only additional significant factor, UGT2B15 genotype, was considered simultaneously (p < 0.001; Table 1). In contrast, neither gender nor ethnicity was significant. Among the fetal samples (N = 71), the continuous variable, increasing gestational age, was the only factor significantly associated with greater UGT2B15 expression (p < 0.001, r2 = 0.621; stepwise linear regression), whereas genotype, gender, and ethnicity were not significant. Among the 165 postnatal samples, the relationship between age categories and greater UGT2B15 expression levels remained the most significant variable when genotype and male gender were considered concurrently (p < 0.001, <0.01, and <0.05, respectively, stepwise linear regression; Table 2). Of note, the effect size for age was about 2- to 3-fold greater than that of genotype or gender (Table 2). With these variables considered, ethnicity was not significantly associated with differences in postnatal UGT2B15 protein content.
Factors Associated With Greater Log-Transformed UGT2B15 Protein Content in 236 Human Hepatic Microsomal Samples. Other Variables Tested Included Gender, Ethnicity, and g.23490A>C Genotype. (Stepwise Linear Regression, Model r2 = 0.687)
| Factor . | Standardized β . | t . | p-value . |
|---|---|---|---|
| Age category | 0.801 | 18.355 | <0.001 |
| Variant Genotype (g.253G/T) | 0.114 | 2.617 | <0.05 |
| Factor . | Standardized β . | t . | p-value . |
|---|---|---|---|
| Age category | 0.801 | 18.355 | <0.001 |
| Variant Genotype (g.253G/T) | 0.114 | 2.617 | <0.05 |
| Factor . | Standardized β . | t . | p-value . |
|---|---|---|---|
| Age category | 0.801 | 18.355 | <0.001 |
| Variant Genotype (g.253G/T) | 0.114 | 2.617 | <0.05 |
| Factor . | Standardized β . | t . | p-value . |
|---|---|---|---|
| Age category | 0.801 | 18.355 | <0.001 |
| Variant Genotype (g.253G/T) | 0.114 | 2.617 | <0.05 |
Factors Associated With Greater Log-Transformed UGT2B15 Protein Content in Postnatal Human Liver Microsomal Samples (n = 165). Other Variables Tested Included Ethnicity. (Stepwise Linear Regression; Model r2 = 0.315)
| Factor . | Standardized β . | t . | p-value . |
|---|---|---|---|
| Increasing postnatal age group | 0.506 | 7.121 | <0.001 |
| Variant Genotype (g.253G/T) | 0.197 | 2.770 | <0.01 |
| Male gender | 0.150 | 2.105 | <0.05 |
| Factor . | Standardized β . | t . | p-value . |
|---|---|---|---|
| Increasing postnatal age group | 0.506 | 7.121 | <0.001 |
| Variant Genotype (g.253G/T) | 0.197 | 2.770 | <0.01 |
| Male gender | 0.150 | 2.105 | <0.05 |
| Factor . | Standardized β . | t . | p-value . |
|---|---|---|---|
| Increasing postnatal age group | 0.506 | 7.121 | <0.001 |
| Variant Genotype (g.253G/T) | 0.197 | 2.770 | <0.01 |
| Male gender | 0.150 | 2.105 | <0.05 |
| Factor . | Standardized β . | t . | p-value . |
|---|---|---|---|
| Increasing postnatal age group | 0.506 | 7.121 | <0.001 |
| Variant Genotype (g.253G/T) | 0.197 | 2.770 | <0.01 |
| Male gender | 0.150 | 2.105 | <0.05 |
To investigate whether gestational age was associated with UGT2B15 content among infants born prematurely, the subset of postnatal samples with known gestational and postnatal ages (N = 29) were analyzed. Among this small sample of infants (postnatal age range: 1–169 days; gestational age range: 21–42 weeks), after adjusting for the previously identified factors of gender and genotype, both postnatal age group and increasing gestational age were modestly associated with greater UGT2B15 expression (p < 0.05, stepwise linear regression, Table 3).
Factors Associated With Greater Log-Transformed UGT2B15 Protein Content in the Subset of 29 Postnatal Human Liver Samples With Known Gestational and Postnatal Age. Other Variables Tested Included Genotype and Gender
| Factor . | Standardized β . | t . | p-value . |
|---|---|---|---|
| Increasing postnatal age group | 0.448 | 2.724 | <0.05 |
| Increasing gestational age | 0.402 | 2.507 | <0.05 |
| Factor . | Standardized β . | t . | p-value . |
|---|---|---|---|
| Increasing postnatal age group | 0.448 | 2.724 | <0.05 |
| Increasing gestational age | 0.402 | 2.507 | <0.05 |
| Factor . | Standardized β . | t . | p-value . |
|---|---|---|---|
| Increasing postnatal age group | 0.448 | 2.724 | <0.05 |
| Increasing gestational age | 0.402 | 2.507 | <0.05 |
| Factor . | Standardized β . | t . | p-value . |
|---|---|---|---|
| Increasing postnatal age group | 0.448 | 2.724 | <0.05 |
| Increasing gestational age | 0.402 | 2.507 | <0.05 |
DISCUSSION
Our study is the first to characterize comprehensively the human hepatic UGT2B15 developmental expression pattern. Other than one 24 week gestation sample, UGT2B15 expression was not observed until the third trimester; only about 80% of that group demonstrated expression and values were six-fold lower than mature samples. UGT2B15 content during late gestation was commensurate with the first weeks of life, suggesting that birth is not an immediate, acute stimulant for expression. Nor did birth appear sufficient as expression was not detected in some neonatal samples. The maturational process was impacted by both prematurity and chronologic age in a small subset of postnatal samples.
Significantly higher UGT2B15-specific content was observed during later infancy and childhood compared with the first three postnatal weeks. The specific mechanism underlying these changes is unknown; however, we speculate that similar to other UGTs, developmental expression involves regulation by liver enriched transcription factors. Specifically, HNF1α, C/EBPα, C/EBPβ, and PAR have been shown to contribute to age-related changes in the hepatic expression of other drug metabolizing enzymes (Cereghini, 1996; Klick et al., 2008; Martinez-Jimenez et al., 2005; Ourlin et al., 1997). Also, several UGT1 and UGT2 genes are variably regulated by HNF1α and HNF4α (reviewed in MacKenzie et al., 2010). However, for UGT2B15, the HNF1α-mediated increase in endogenous expression required trichostatin A, a histone deacetylase inhibitor (MacKenzie et al., 2010), suggestive that chromatin remodeling may also be important. The UGT2B15 promoter has been shown to bind HNF4α, a zinc-finger nuclear receptor, but binding did not alter UGT2B15 transcription (MacKenzie et al., 2010; Odom et al., 2004). Although functionally untested, several other transcription factors, including HNF3β, HNF3γ, C/EBPα, and C/EBPβ, have recognition sites within 300 bases from the ATG translation start site (Transfac, http://transfac.gbf.de). Thus, it is plausible that these liver-enriched transcription factors could contribute to UGT2B15 age-related expression changes.
Previously, our knowledge of human UGT2B15 ontogeny was based solely on human hepatic mRNA data, a poor proxy for protein expression (Ekstrom et al., 2013; Nahar et al., 2013; Strassburg et al., 2002). Prior reports were limited by small sample sizes and/or the studies’ restriction to relatively early life and adulthood, without intervening sampling times. Nevertheless, consistent with our data, Ekstrom et al. (2013) reported a 36-fold greater UGT2B15 mRNA expression in adult versus fetal livers. Court et al. (2012) detected transcripts in 63 pooled fetal livers ranging from 22 to 40 weeks gestation. UGT2B15 transcripts were not detected in two fetal livers at 20 weeks gestation by Strassburg et al. (2002), but adult transcript levels were detected by seven months of age (N = 28 postnatal samples). In contrast, two studies detected UGT2B15-specific mRNA in 50 or more fetal livers prior to 12 (Ekstrom et al., 2013) and 17 weeks (Nahar et al., 2013). However, the problem with using mRNA, as done in all these studies, was demonstrated by incongruent transcript and protein levels in human embryonic kidney 293—UGT2B15 stably transfected cells (Turgeon et al., 2001). This disparity suggests that post-transcriptional processing, translational efficiency, and post-translational modifications alter protein levels. Thus, the incongruity between our data and the prior reports of mRNA is not surprising.
The direct quantitative hepatic UGT2B15 protein measurements herein fill an important gap for physiologically based pharmacokinetic modeling for human risk assessment. This approach predicts the disposition of compounds that cannot be ethically given to pregnant women, infants, and children directly (e.g., Nong et al., 2006). This modeling method has been used to evaluate the UGT2B15 substrate, BPA in adults (Teeguarden et al., 2005). However, the unavailability of UGT2B15 protein expression data during development precluded reliably estimating age-dependent BPA pharmacokinetic differences among infants, children, and adults using these models. BPA, used in the manufacture of polycarbonate plastics and epoxy resins used in many consumer products, attracted national attention because of widespread exposure and potential toxicity, believed to be limited to its unconjugated form (Richter et al., 2007). Greater BPA concentrations present in younger animals, compared with older animals dosed similarly, have been presumed to be secondary to decreased conjugation (Domoradzki et al., 2004). Yet, developmental changes affecting human BPA metabolism have remained unclear. Thus, the human hepatic UGT2B15 protein values generated herein are a substantive addition to improve BPA internal dosimetry modeling.
The expression pattern we observed, that is hepatic UGT2B15 protein levels at about 18% of mature values during the second and third trimester with maturation by 15 weeks postnatal age, is consistent with other UGTs, closely resembling other UGT2B family members. UGT2B7 and UGT2B17 expression onset is detectable in second trimester fetal livers at 10–20% and 3% of adult values, respectively (Leakey et al., 1987; Pacifici et al., 1982). UGT2B7 adult levels are attained by two to six months, whereas the maturation time for UGT2B17 remains unknown (Anderson et al., 1997; Leakey et al., 1987; Pacifici et al., 1982). Limited UGT2B4 expression data indicate that the transcript levels in pediatric livers, ages 6–24 months, were 30–40% of adult levels (Strassburg et al., 2002). Among the UGT1A members, UGT1A1, the enzyme predominantly responsible for bilirubin glucuronidation, is similar to UGT2B15, being non-detectable in livers between 16 and 25 weeks gestation, but with adult levels attained by three to six months postnatal age (Burchell et al., 1989). In contrast, UGT1A6 and 1A3 expression onset occurs in the fetal liver, but at only about 10% and 30% of adult expression, respectively (Burchell et al., 1989; Coughtrie et al., 1988). UGT1A6 activity reaches approximately 50% maturity by six months, with full maturation occurring at puberty (Alam et al., 1977; Coughtrie et al., 1988). The complete UGT1A3 ontogeny remains uncharacterized (Burchell et al., 1989) (reviewed in de Wildt et al., 1999). In a study of only postnatal liver, hepatic UGT1A4 maturation was reported to occur by 1.4 years of age (Miyagi and Collier, 2007). UGT1A9 protein expression and activity were not detected at birth, but age-dependent expression matured by about four months (Miyagi et al., 2012). In summary, age is a determinant of human hepatic UGT protein expression, with some UGTs, particularly those in the UGT2B family having a similar maturational pattern to that observed for UGT2B15.
We found a 31-fold variation in UGT2B15-specific content across the entire sample, which is comparable to the 21-fold variability seen with UGT2B15-mediated oxazepam glucuronidation (Court et al., 2004). Prior studies found that the UGT2B15 g.253G>T SNP is associated with decreased protein (Court et al., 2002; Levesque et al., 1997). In our study, the g.253G>T variant was associated with significantly greater UGT2B15 protein expression in the postnatal samples. The discrepancy between the present study and the prior in vitro reports may be partially explained by the microsomal source. We utilized human hepatic microsomes, whereas others used stably transfected human embryonic kidney cell line derived microsomal fractions (Court et al., 2002; Levesque et al., 1997). Further, Court et al. speculated that the differences in plasmid copy number between UGT2B15 *1 and *2 stably transfected clones may have impacted protein expression differences (Court et al., 2002). Thus, the direction of the genotype effect on expression is unclear. In addition to altering UGT2B15 protein expression, the g.253G>T SNP has been associated with both increased and decreased enzyme activity in a substrate-specific fashion (Court et al., 2002; Levesque et al., 1997). Irrespective of the directional effect, given that half the population has at least one copy of the g.253T variant, the functional effect we observed supports further evaluation of this SNP within human in vivo disposition studies of compounds such as BPA.
In postnatal subjects, males had higher UGT2B15-specific content, even after adjustment for age and genotype. Most, but not all, authors have shown similar findings using both in vitro and in vivo approaches. The median S-oxazepam glucuronidation was 67% greater among male livers (Court et al., 2004), and mean oxazepam clearance was 40% greater among men (Greenblatt et al., 1980). Gender differences in BPA disposition have been reported, with men demonstrating a twofold greater BPA glucuronidation compared with women (Kim et al., 2003). In contrast, Gallagher et al. (2010) did not observe any UGT2B15 mRNA expression level differences between genders among adult liver samples obtained from elderly hepatocellular carcinoma patients. This gender difference in hepatic UGT2B15 expression and activity is interesting, as the enzyme inactivates male steroid hormones, including testosterone and dihydrotestosterone (Court et al., 2004; Levesque et al., 1997).
Developmental changes in protein expression can lead to an increased risk of therapeutic failure and adverse drug reactions in the fetus and children (reviewed in Hines, 2008). The present study defined UGT2B15 ontogeny in a developmentally diverse set of liver samples by quantitating the hepatic protein content. We did so while simultaneously considering genotype, ethnicity, and gender as determinants of UGT2B15 protein expression. Protein absence during early gestation with relatively low expression during late gestation and the neonatal period are consistent with greater risks from some UGT2B15 substrates during this time window. For example, given the role of UGT2B15 in BPA metabolism, developmental expression may have a substantial effect on its disposition in the very young.
FUNDING
Public Health Service grant (5R01ES017764); Children's Research Institute, Children's Hospital of Wisconsin, Milwaukee, WI.
The authors would like to thank Dr Pippa Simpson for her insightful discussion about statistical analyses.
Current address: U.S. Environmental Protection Agency, Office of Research and Development, National Health and Environmental Effects Research Laboratory, 109 T.W. Alexander Dr., Research Triangle Park, NC 27711.
REFERENCE

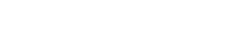


Comments