1
Howard Hughes Medical Institute, Department of Biological Sciences, Columbia University, New York, NY, USA
2
Departamento de Química Inorgánica Analítica y Química Física, Facultad de Ciencias Exactas y Naturales, Universidad de Buenos Aires, Buenos Aires, Argentina
We describe neurobiological applications of RuBi-Glutamate, a novel caged-glutamate compound based on ruthenium photochemistry. RuBi-Glutamate can be excited with visible wavelengths and releases glutamate after one- or two-photon excitation. It has high quantum efficiency and can be used at low concentrations, partly avoiding the blockade of GABAergic transmission present with other caged compounds. Two-photon uncaging of RuBi-Glutamate has a high spatial resolution and generates excitatory responses in individual dendritic spines with physiological kinetics. With laser beam multiplexing, two-photon RuBi-Glutamate uncaging can also be used to depolarize and fire pyramidal neurons with single-cell resolution. RuBi-Glutamate therefore enables the photoactivation of neuronal dendrites and circuits with visible or two-photon light sources, achieving single cell, or even single spine, precision.
Caged compounds are becoming widely used in neuroscience because they enable the optical manipulation of neuronal circuits (Hess, 1999
). Using photoreleasable neurotransmitters such as glutamate and GABA, one can activate or inhibit neuronal processes, single neurons, or groups of neurons (Ellis-Davies, 2005
). Combining this with functional imaging provides a near ideal method of monitoring and controlling neuronal activity, allowing an entirely optical method for analysis of neural circuits (Nikolenko et al., 2007
).
In particular, two-photon uncaging of glutamate, with its excellent spatial resolution, has been quite useful, due to the relatively high two-photon absorption cross sections of functionalized nitrobenzyl derivatives, such as 7-nitroindolinyl- and 4-methoxy-7-nitroindolinyl-amino (MNI) glutamate (Canepari et al., 2001
; Matsuzaki et al., 2001
). For example, two-photon uncaging of MNI-glutamate has been used successfully to functionally map synaptic receptors (Matsuzaki et al., 2001
), activate individual spines (Araya et al., 2006
; Carter and Sabatini, 2004
; Gasparini and Magee, 2006
; Matsuzaki et al., 2004
; Sobczyk et al., 2005
) and activate individual neurons (Nikolenko et al., 2007
). In spite of its utility, MNI-glutamate needs to be applied to the tissue at relatively high (mM) concentrations for effective two-photon uncaging. Unfortunately, at these concentrations, MNI-glutamate, like other caged compounds, is a very effective antagonist of GABAergic transmission (see below for MNI glutamate; Maier et al., 2005
; Molnar and Nadler, 2000
for other compounds). This can be a significant problem for its use in the study of neuronal circuits, as its application can lead to epileptiform events. Also, as it strongly blocks the GABAergic responses, it prevents the study of inhibitory network activity or connectivity.
To help circumvent this problem, we have exploited the flexibility of ruthenium chemistry, and have synthesized a novel series of caged compounds, using ruthenium-bipyridine complexes (Zayat et al., 2003
, 2005
). Ruthenium is a transition metal, widely used due to its versatile chemistry. The polypyridines of ruthenium photorelease entire ligands in a heterolytic fashion, by means of a widely known mechanism in which the initial photoexcited state quickly evolves into a dissociative state, so the photorelease is therefore clean and fast (Zayat et al., 2003
). We recently introduced a caged GABA compound (“RuBi-GABA”) that can be used for optical inactivation of neurons and functional mapping of GABAergic currents (Rial Verde et al., 2008
). Now we describe the biological characterization of a new caged glutamate compound, “RuBi-Glutamate”, which can be photoreleased with one- or two-photon excitation and has less nonspecific effects than MNI glutamate. The chemical synthesis and photochemical characterization of RuBi-Glutamate is described elsewhere (Salierno et al., submitted
). Although we expect that RuBi-Glutamate will be useful for one-photon visible uncaging experiments, in this report we mainly focus in its use for two-photon photostimulation, since there are already several adequate choices available for neuronal photostimulation with one-photon uncaging of glutamate (Callaway and Katz, 1993
; Callaway and Yuste, 2002
; Frick et al., 2001
; Pettit et al., 1997
; Shepherd et al., 2003
; Wieboldt et al., 1994
; Yoshimura et al., 2005
).
Slice Preparation and Electrophysiology
350 μm thick coronal slices from 14-day-old C57BL/6 mouse cortex were prepared using a Leica VT1000-S vibratome with a cutting solution containing (in mM): 27 NaHCO3, 1.5 NaH2PO4, 222 Sucrose, 2.6 KCl, 2 MgSO4, 2 CaCl2. Slices were incubated at 32°C in ACSF for 30 min and then kept at room temperature for at least 30 min before transferring them to the recording chamber. The recording chamber was bathed in ACSF (pH 7.4), also kept at room temperature and saturated with 95% O2 and 5% CO2, containing (in mM): 126 NaCl, 3 KCl, 2 MgSO4, 2 CaCl2, 1 NaH2PO4, 26 NaHCO3, and 10 glucose.
Neurons were either held at their resting membrane potential (−65 mV), or at +40 mV, when both inhibitory and excitatory events were recorded. Whole-cell electrodes (4–7 MΩ) were used. To establish whole-cell access the cells were illuminated with an oblique light and an IR-pass filter on the microscope field diaphragm, and visualized through a CCD camera (DAGE-MTI IR-1000) connected to a Sony PVM-137 black and white video monitor. All recordings, except those shown in Figures 2 and 3
were performed with intracellular solution (pH 7.25), containing (in mM): 135 K-methylsulfate, 10 KCl, 10 HEPES, 5 NaCl, 2.5 Mg-ATP, 0.3 Na-GTP, and 0.1 Alexa Fluor 594. Voltage-clamp recordings shown in Figures 2 and 3
were done with intracellular solution (pH 7.25), containing (in mM): 128 Cs-methanesulfonate, 10 HEPES, 2 MgCl2, 2 MgSO4, 4 Na2-ATP, 0.4 Na-GTP, 10 Na2-phosphocreatine and 0.1 Alexa Fluor 594. Experiments were conducted at room temperature (22–25°C). We performed recordings from layer 2/3 and 5 pyramidal cells using MultiClamp 700B (Molecular Devices) amplifiers, and acquired the signals through a National Instruments PCI 6259 board using either custom software developed with LABView or Matlab. In a subset of experiments, electrical stimulations were performed using a bipolar electrode placed in layer 5. Repetitive stimuli were applied at a frequency of 0.2 Hz. CNQX (20 μM) and APV (40 μM) were routinely used while recording IPSCs, and TTX (2 μM) was added to the ACSF to isolate miniature synaptic currents.
Off-line analysis was conducted using Matlab, MiniAnalysis v6.0.7 or IGOR Pro with the Neuromatic v2.0 package. For consistency throughout the study, all measurements are expressed as mean ± SEM. Statistical significance was assessed using Student’s t-test and the non-parametric Mann–Whitney test when appropriate at the significance level (p) indicated.
RuBi-Glutamate Uncaging
Images were acquired using a custom-made two-photon laser scanning microscope based on the Olympus FV-200 system (side-mounted to a BX50WI microscope with a 40× , 0.8NA, water immersion objective) and a Ti:sapphire laser (Chameleon Ultra II, Coherent, >3 W, 140 fs pulses, 80 MHz repetition rate). Fluorescence was detected with a photomultiplier tube (PMT: H7422-P40 Hamamatsu, Bridgewater, NJ, USA) connected to a signal amplifier (Signal Recovery AMETEK Advanced Measurement Technology, Wokingham, UK) whose output was connected to the Fluoview system. First, images of the soma were acquired with Fluoview software (XY scan mode with 1× to 10× digital zoom), at 850 nm using minimal power to prevent RuBi-Glutamate uncaging. As RuBi-Glutamate is light-sensitive, the computer screens and video monitor were covered with two layers of Rosco #27 medium red filters.
A somatic uncaging point was selected using custom software (Nikolenko et al., 2007
), which triggered the uncaging pulse and controlled the pulse duration. Laser power was modulated by a Pockels cell (Quantum Technology, Lake Mary, FL, USA). For somatic stimulations, each uncaging target consisted of eight subtargets, each of which was illuminated for 8 ms, giving a total duration of ∼70 ms. The subtargets themselves were complex, consisting of five very closely spaced beamlets created by multiplexing the laser beam with a diffractive optical element (DOE). The stimulations were repeated every 4 s. RuBi-Glutamate was added to the bath at 300 μM concentration. The concentration chosen for two-photon experiments, 300 μM, was the lowest concentration with which we were able to fire reliably using our stimulation protocol. While we performed other experiments with lower concentrations of RuBi-Glutamate (∼150 μM), neurons did not fire consistently (or sometimes not at all). One-photon experiments, were performed with 30 μM the RuBi-Glutamate.
A Dynamax peristaltic pump (Ranin Instruments Inc., Woburn, MA, USA) was used to control bath perfusion to minimize total bath volume and re-circulate oxygenated media. Uncaging was performed with 800 nm light for two-photon experiments, and with 473 nm light for one-photon experiments.
To measure the lateral two-photon uncaging spatial resolution in single spines, the laser beam was positioned near the spine head (∼0.2 μm from the spine head edge) and subsequently moved away from the spine head edge. We specifically decided not to uncage glutamate directly over the spine head to avoid damaging the spine membrane, which we know can be harmed by direct laser illumination. Instead, to prevent photodamage and trigger reliable glutamate responses without affecting the spine head, we directed the beam right next (∼0.2 μm) from the spine head edge. With this stimulus protocol, we can reliably photostimulate spines for long periods of time without any appreciable photodamage. In addition, to measure the axial resolution of the RuBi-Glutamate uncaging responses, the laser beam was positioned next to the spine head and the uncaging responses were recorded at different focal planes (spaced every 0.8 μm). In these tests, 4 ms laser pulses at 2 s interval were used. To record desynchronized spontaneous miniature excitatory post-synaptic potentials (mEPSPs) from layer 5 neurons, we replaced extracellular calcium with strontium (Xu-Friedman and Regehr, 2000
). Comparisons between mini-PSPs (mPSPs) or mini-PSCs (mPSCs) and photo-evoked responses are confounded by the spatial filtering. mPSPs or PSCs distributions are broad, stemming in part from the fact that the events are arising at many different spatial locations, each with a different cable-length filtering to the recording electrode, whereas in our photo-evoked responses the distances are much more uniform.
Two-Photon Uncaging of RuBi-Glutamate Activates Glutamate Receptors
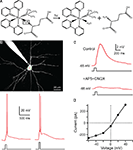
RuBi-Glutamate (cis-[Ru(bpy)2(PMe3)GluH2](PF6)2, with bpy = 2,2′bipyridine and PMe3 = trimethylphosphine) is a caged glutamate compound composed of a ruthenium center with six coordination positions, four of which are occupied by the two bidentate bipyiridines, the fifth by a trimethylphosphine and the sixth by a molecule of L-glutamate, coordinated through its amino group (Figure 1
A). Upon absorption of one photon of visible light, or two near infrared photons, RuBi-Glutamate photocleaves within nanoseconds, and releases glutamate from the Ru-bipyridine core, with a quantum yield of ∼0.13 at pH 7, and an extinction coefficient of >4,000 M−1 cm−1 at 473 nm (Salierno et al., submitted
).
Figure 1. RuBi-Glutamate uncaging activates glutamate receptors. (A) Structure of Ruthenium-bipyridine-trimethylphosphine-Glutamate (RuBi-Glutamate) and glutamate photorelease reaction. (B) Upper panel: Layer 2/3 pyramidal cell loaded with Alexa-594 and position of the multiplexed uncaging laser targets (eight subtargets) on the soma of the cell. Lower panel: Action potentials triggered by uncaging of RuBi-Glutamate (300 μM) in a layer 2/3 pyramidal neuron. In this example, one action potential was evoked with a power on sample of 180 mW and two with 220 mW. (C) RuBi-Glutamate activates glutamate receptors. Adding APV (40 μM) and CNQX (20 μM) blocked the uncaging response. (D) Representative current–voltage plot of uncaging responses. Note how it reverses at +10 mV, indicating that the responses are mediated by glutamate receptors.
We first examined the ability of RuBi-Glutamate to activate glutamate receptors by performing whole-cell recordings from layer 2/3 pyramidal neurons in slices from mouse neocortex. Slices were bathed with a solution containing the caged form of RuBi-Glutamate and the laser beam was aimed toward the somata of the recorded neurons, uncaging glutamate, while simultaneously recording the membrane potential from the activated neuron, and monitoring the effect of the uncaging. Using this functional assay to probe two-photon uncaging, we explored several different excitation wavelengths and concentrations and found 800 nm to be optimal and 300 μM a sufficient RuBi-Glutamate concentration to generate reliable functional responses. For the one-photon studies mentioned here, we used 473 nm light and 30 μM RuBi-Glutamate, although because of the high absorption cross section for single photon excitation and the larger one-photon point spread function (PSF), we were also able to generate responses with concentrations as low as ∼5 μM (data not shown). These wavelengths and concentrations agree with those expected by examining the photochemical properties of RuBi-Glutamate (Salierno et al., submitted
), and also match those previously used in our work with other members of that family of RuBi-caged compounds, such as RuBi-GABA (Rial Verde et al., 2008
).
In pyramidal neurons, recorded under current clamp, two-photon uncaging of glutamate generated brief depolarizations, but with a single laser spot, the evoked depolarizations were too small to reliably cause the cell to fire action potentials. We therefore used a DOE to spatially multiplex the laser beam into five closely spaced beamlets, a strategy that we have previously introduced with MNI glutamate photostimulation (Nikolenko et al., 2007
). This beamlet array was then directed onto eight subtargets on the cell (Figure 1
B, upper panel), releasing more glutamate, generating stronger cellular responses leading to action potentials. Using this protocol, two-photon uncaging (on average, ∼70 ms, 150–400 mW on the sample; n = 15 neurons) was able to reliably generate action potentials (Figure 1
B, lower panel). Uncaging depolarizations ranged in amplitude from 1 to 20 mV and were effectively and reversibly blocked by bath application of the glutamate receptor antagonists APV/CNQX (Figure 1
C; 40/20 μM respectively; 96.7 ± 1.5% reduction in voltage, n = 6 neurons). In addition, in voltage-clamp conditions, RuBi-Glutamate uncaging generated inward currents that reversed at +10 mV, indicative of a glutamate receptor current (Figure 1
D). These data confirmed that the uncaging of RuBi-Glutamate activated glutamate receptors in cortical pyramidal neurons.
Effect of RuBi-Glutamate on Spontaneous PSCs
We then characterized the pharmacological effect of RuBi-Glutamate on synaptic receptors. As mentioned, the near complete blockade of inhibitory synaptic transmission by MNI-glutamate at concentrations useful for two-photon excitation (see below, Figure 3
) prevents its clean use for circuit studies. To examine the effects from RuBi-Glutamate, we performed voltage-clamp recordings of spontaneous synaptic currents at different holding potentials, before and after bath application of RuBi-Glutamate. We performed these experiments with 300 μM, the same concentration of RuBi-Glutamate that we used for two-photon uncaging. We also carried out experiments in the presence and absence of the sodium channel blocker TTX, in order to specifically test whether mPSCs were specifically affected by RuBi-Glutamate.
At resting membrane potential of −65 mV, bath application of 300 μM RuBi-Glutamate had no detectable effect on the average amplitude, frequency or rise time of spontaneous PSCs or of mPSCs in the presence of TTX (Figure 2
A, n = 1,562 events from six control neurons, 1,775 from six neurons in the presence of RuBi-Glutamate). Since at −65 mV most PSCs are excitatory, these data indicated that bath applied RuBi-Glutamate had no significant effect on spontaneous excitatory PSCs (EPSCs). We then examined its potential effect on inhibitory PSCs (IPSCs), by performing similar experiments at +40 mV holding potential, a level at which both excitatory and inhibitory PSCs can be readily detected. At +40 mV, bath application of 300 μM RuBi-Glutamate produced a general trend of reducing the amplitude and frequency of spontaneous and miniature PSCs, with no apparent changes in the rise time of the events (Figure 2
B). Although these effects were not statistically significant, in the presence of TTX, we did observe a significant reduction in PSCs frequency (p < 0.001, n = 6, t-test). This result, together with the lack of significant effect at −65 mV, close to the reversal potential of IPSCs, indicated a potential antagonistic effect of the caged form of RuBi-Glutamate on inhibitory currents.
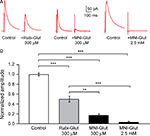
Figure 2. Effect of RuBi-Glutamate on spontaneous synaptic currents. (A) Effects of RuBi-Glutamate on spontaneous activity recorded at a holding membrane potential of −65 mV. At this resting potential, RuBi-Glutamate (300 μM) had no significant effect on the average amplitude, frequency or rise time of spontaneous PSCs (n = 6 neurons) or of miniature PSCs recorded in the presence of TTX (n = 6 neurons). (B) Effects of RuBi-Glutamate on spontaneous activity recorded at +40 mV. RuBi-Glutamate (300 μM) tends to reduce the amplitude and frequency of spontaneous and miniature PSCs, but only the reduction of frequency of miniature PSCs is statistically significant (p < 0.001, n = 6 neurons). There was no change in the rise time of the events. Because both excitatory and inhibitory events are recorded at +40 mV, this indicates that RuBi-Glutamate may have an inhibitory effect on IPSCs. Error bars represent SEM. ***p < 0.001.
Effect of RuBi-Glutamate on Evoked Inhibitory Transmission
To directly examine if RuBi-Glutamate had an antagonistic effect on IPSCs we conducted a series of experiments in which we evoked inhibitory currents by stimulating the neuropil surrounding the recorded neuron with a stimulating electrode, while holding the neuron at +40 mV, in the presence of APV and CNQX (40/20 μM, respectively) and thus better isolate IPSCs in the presence or absence of RuBi-Glutamate. The rationale of these experiments was to test whether RuBi-Glutamate blocked physiological synaptic GABAergic transmission, since our ultimate aim was to use two-photon uncaging of RuBi-Glutamate for circuit studies. Side-by-side experiments were performed with MNI-glutamate in order to examine its effect on GABAergic currents. We tested the effects from MNI-Glutamate at both 300 μM, the concentration used in our RuBi-Glutamate uncaging experiments, and at the higher concentrations of MNI-Glutamate needed (>2.5 mM), in our experience, for effective two-photon MNI-glutamate uncaging.
In these experiments, the perfusion of 300 μM RuBi-Glutamate generated a significant reduction of the peak amplitude of the evoked GABAergic IPSCs (Figure 3
: 50.0 ± 6.0%, n = 7 cells, p < 0.001). Perfusion of MNI-glutamate at the same concentrations (300 μM) generated an even more pronounced decrease in evoked IPSCs (Figure 3
B; 83.0 ± 3.0%, n = 4, p < 0.001). At 2.5 mM, the concentration needed for effective two-photon uncaging, MNI-glutamate completely blocked the evoked response (Figure 3
: 97.0 ± 2.0%, n = 6, p < 0.001).
Figure 3. Effect of RuBi-Glutamate and MNI-glutamate on inhibitory transmission. (A) Voltage-clamp responses of a pyramidal neuron to extracellular electrical stimulation. An external bipolar stimulation electrode was used to evoke responses while clamping the neuron at +40 mV and in presence of APV (40 μM) and CNQX (20 μM) in the ACSF to isolate the GABA currents. Here we show examples of IPSCs evoked in control conditions, and the diminution of their amplitude after adding either RuBi-Glutamate (300 μM, left part) or MNI-Glutamate (300 μM or 2.5 mM, right part) to the bath. (B) Histogram showing averages of the inhibition of GABA current in presence of RuBi-Glutamate or MNI-Glutamate. **p < 0.01, ***p < 0.001.
We concluded that, although 300 μM RuBi-Glutamate has a significant antagonist effect on endogenous GABAergic currents, this effect (close to 50%) is smaller compared with that of MNI glutamate (∼80% at 300 μM, ∼100% at the effective concentration of 2.5 mM). The differences between the 50% reduction observed in these RuBi-Glutamate experiments vs. the smaller reduction observed in the +40 mV recordings of spontaneous miniature potentials in TTX (Figure 2
) may be partly due to the fact that at +40 mV one records a mixture of EPSCs and IPSCs.
RuBi-Glutamate Uncaging can Activate Individual Dendritic Spines
We then tested the use of two-photon uncaging of RuBi-Glutamate for activating dendritic spines, one of the most useful applications of two-photon glutamate uncaging (Araya et al., 2006
; Carter and Sabatini, 2004
; Gasparini and Magee, 2006
; Matsuzaki et al., 2004
; Sobczyk et al., 2005
). For these experiments, we performed whole-cell recordings from pyramidal neurons and filled them with Alexa 594 to optimally visualize their dendritic spines. We then bathed the slice in RuBi-Glutamate (300 μM) and directed a pulsed laser at the neuropil adjacent to a chosen spine (Figures 4
A,B). With 4 ms laser pulses, we triggered reliable depolarizations in the neuron (see traces in Figures 4
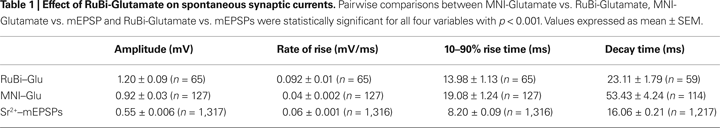
C,D). These responses were similar in amplitude to events recorded after two-photon uncaging of MNI-glutamate (see Table 1
). However, the RuBi-Glutamate generated neuronal responses had a rate of rise nearly twice as fast as those generated from uncaging MNI-glutamate (p < 0.001, n = 65 RuBi-Glutamate uncaging events from four spines vs. n = 127 MNI-glutamate uncaging events from nine spines). In addition, 10–90% rise times and 37% decay time kinetics were faster then those observed with uncaging of MNI-glutamate (p = 0.001 and p < 0.001, respectively, Table 1
), and more closely resemble the kinetics of spontaneous mEPSP (Table 1
, see Materials and Methods for detection of mEPSPs).
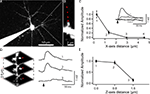
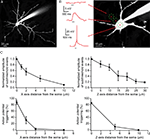
Figure 4. Use of RuBi-Glutamate uncaging to optically activate dendritic spines. (A) Layer 2/3 neuron loaded with Alexa-594 to detect dendritic spines. (B) Higher resolution image of a dendritic spine selected for uncaging. Red dots indicate the sites of uncaging. (C) Plot of the XY spatial resolution of uncaging from the spine shown in (B) (Inset, traces corresponded to averages of 15 uncaging potentials at the different locations [1–4; red dots in (B)]. (D) Spatial axial resolution of uncaging. Z-stack of images from the stimulated spine. Traces corresponded to averages of 15 uncaging potentials of its corresponding image stack. Red dots indicate the site of laser beam parking, not the actual size of the beam profile. (E) Plot of the axial resolution at the three different locations showed in (D).
In these single spine experiments, we also explored the spatial resolution of the uncaging responses, by monitoring the evoked depolarization while changing the position of the laser with respect to the dendritic spine. Moving the uncaging spot perpendicular to the spine (i.e., in the XY plane) resulted in a strong decrease in the amplitude of the uncaging response (∼80% reduction with the beam 1.3 μm away from the spine, Figure 4
C; n = 7 spines and 2 neurons). In the axial (Z) direction, a small change in position, 0.8 μm, also resulted in a strongly diminished voltage response (Figures 4
D,E; 71% average reduction; n = 30 uncaging events, n = 2 neurons). These results are consistent with the small local uncaging volume defined by the two-photon PSF, and are similar to measurements obtained with two-photon uncaging of MNI-glutamate (Araya et al., 2006
). Thus, two-photon uncaging of RuBi-Glutamate has a spatial resolution that approximates the dimension of a typical dendritic spine, and therefore can be used to selectively activate individual spines.
RuBi-Glutamate Uncaging can Activate Individual Neurons
Finally, we tested the potential use of RuBi-Glutamate for the optical photostimulation of neurons. These types of experiments, in which one uncages glutamate to elicit action potentials from a desired neuronal target, have been very useful for mapping the synaptic inputs in slices (Callaway and Katz, 1993
; Shepherd et al., 2003
; Yoshimura et al., 2005
). Moreover, with two-photon excitation, these inputs maps can be built with single-cell precision (Nikolenko et al., 2007
).
These experiments were performed by recording from pyramidal neurons with whole-cell electrodes and directed the uncaging laser onto the soma of the cells, in slices bathed with 300 μM RuBi-Glutamate (Figure 5
A). As previously mentioned, we used eight subtargets and a DOE to spatially multiplex the laser beam (Figure 5
B). With this protocol we reliably generated action potentials in the recorded neuron (90.0 ± 2.6% action potential occurrence, ∼70 ms uncaging pulses, n = 11 cells; Figure 5
).
Figure 5. Use of RuBi-Glutamate uncaging to optically activate neurons. (A) Layer 2/3 pyramidal cells were filled with Alexa-594. (B) Position of the uncaging targets (eight subtargets). When placed on the somata, the uncaging elicited an action potential. When the targets were moved away from the soma, uncaging elicits only subthreshold depolarizations, and no response was evoked 10 μm away. (C) Lateral (XY plane) and axial (Z plane) resolution of RuBi-Glutamate uncaging to elicit either supra- or subthreshold events. Note how subthreshold depolarizations were strongly reduced when targets were placed 10 μm away from the soma in X-axis and 30 μm away in Z-axis (upper panel). Uncaging did not elicit an action potential if targets were moved from the soma 2 μm away in X-axis and 5 μm away in Z-axis (lower panel).
We characterized the effective spatial resolution of our multispot uncaging protocol by moving the spatiotemporally multiplexed laser pattern away from the somata of the cell (Figure 5
B), and measuring the amplitude of subthreshold postsynaptic depolarizations and the probability of suprathreshold events as a function of the distance of the closest target from the cell edge. For subthreshold events, in the XY plane, we measured a strong decrease in the amplitude of the uncaging response, with a minimal response at 10 μm away from the soma (normalized amplitude: 0.10 ± 0.01, n = 8 neurons; Figure 5
C). Movements in the Z plane resulted in a smaller modulation of the response amplitude (normalized amplitude: 0.19 ± 0.02 at 30 μm, n = 5 neurons; Figure 5
C). Because the multiple beamlets and target pattern are dispersed in space, the subthreshold event spatial resolution was understandably worse than that that is obtained with single laser spots (Figure 4
). Nevertheless, for suprathreshold events, we observed a strong decrease of the occurrence of evoked action potentials as the uncaging targets were moved only 2 μm from the edge of the soma in XY plane (3.0 ± 2.8% of occurrence of an action potential, n = 7 neurons) and 5 μm above the soma in Z plane (10.0 ± 7.6%, n = 4 neurons; Figure 5
C). In these experiments, we avoided uncaging near any prominent dendritic processes. However, the same stimulation conditions used to generate an action potential while targeting the soma, only generate subthreshold responses when intentionally (or accidentally) targeting the dendrites.
We also explored the use of one-photon RuBi-Glutamate uncaging in similar experiments, using 30 μM RuBi-Glutamate and a single laser spot, aimed at the center of the somata. With 1 ms laser pulses, one-photon RuBi-Glutamate uncaging generated reliable depolarizations in the cell. The spatial resolution of this response was different in the XY and Z planes, with minimal responses at 15 (normalized amplitude: 0.14 ± 0.03, n = 4 neurons) and 40 μm (normalized amplitude: 0.20 ± 0.03, n = 3 neurons), respectively.
These experiments demonstrated that RuBi-Glutamate can be used, in both one- and two-photon mode, for reliable photostimulation of neurons in brain slices. In particular, the spatial resolution of two-photon DOE uncaging appears adequate for studies with single-cell precision.
Here we introduce the use of RuBi-Glutamate as a novel chemical tool for the photoactivation of individual dendritic spines and cortical neurons in circuits. In the past, glutamate uncaging has been successfully used to map circuits with one-photon excitation (Bureau et al., 2004
; Callaway and Katz, 1993
; Shepherd et al., 2003
; Yoshimura et al., 2005
). The synthesis of MNI-glutamate (Canepari et al., 2001
; Matsuzaki et al., 2001
) enabled the two-photon photoactivation of spines (Araya et al., 2006
; Matsuzaki et al., 2004
) and neurons (Nikolenko et al., 2007
), although the unspecific effects of MNI-glutamate on synaptic receptors have prevented its use for two-photon studies of inhibitory circuits.
RuBi-Glutamate has a number of desirable properties, such as a relatively high absorption cross section in the visible (blue) and a high quantum efficiency of uncaging (Salierno et al., submitted
). This combination allows RuBi-Glutamate to be used at lower concentrations than MNI-glutamate, lessening the blockade of GABAergic responses. The ability of the RuBi-compounds to absorb and uncage with visible light, compared to other caged glutamate compounds, which require UV excitation, is also significant, as blue light has higher penetration and lower toxicity for living tissue. Although we have only performed a cursory exploration of the use of RuBi-Glutamate for one-photon uncaging, we should mention that it has some useful features that other users may find it potentially interesting for one-photon experiments. Specifically, we have effectively uncaged glutamate with visible light from a variety of sources including standard mercury/xenon lamps, LED sources, or even laser pointers and flashlights, something that makes the instrumentation needed for glutamate uncaging experiments quite accessible and inexpensive. Also inexpensive glass optics instead of UV-transparent quartz can be used for uncaging of RuBi-Glutamate.
Perhaps more importantly, the red-shifted single photon spectrum leads to a red-shifted two-photon uncaging peak (800 nm for RuBi-Glutamate vs. 725 with MNI-Glutamate). In addition to the deeper penetration into tissue, 800 nm is near the peak of the maximum power output for the Ti-Sapphire lasers commonly used for two-photon experiments, which means more power is available to uncage glutamate at multiple sites simultaneously, an important consideration for future experiments aimed at probing complex neural circuit dynamics (Nikolenko et al., 2008
).
Our initial experience with RuBi-Glutamate has been positive and we report a number of advantages that could make it widely used. It is water-soluble and does not have any strong pharmacological effects at the concentrations explored for effective one- or two-photon uncaging. Although at higher concentrations (300 μM) it significantly reduces GABAergic responses, it does not block them, so it could allow for the mapping of inhibitory circuits, and the stimulation of dendritic spines in regimes where inhibition is still partly engaged, resulting in more physiological experiments. This, combined with the decreased phototoxicity and increased penetration depth of the longer wavelengths required for uncaging RuBi-Glutamate compared with the nitrobenzyl and nitroindolinyl glutamate derivatives, could make in vivo uncaging experiments feasible.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
We thank Alan Woodruff for initially characterizing the effects of MNI-glutamate on IPSCs and members of both laboratories for help and comments. Supported by the Kavli Institute for Brain Science, the National Eye Institute, the Fulbright Commission, CONICET, ANPCyT, the Office of Technology Ventures from Columbia University and the “Fondation pour la Recherche Médicale” (FRM).